Search Count: 26
 |
Organism: Gammaproteobacteria
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: URE |
 |
Organism: Gammaproteobacteria, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: DNA BINDING PROTEIN Ligands: MN, DCP |
 |
Organism: Yasminevirus sp. gu-2018
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-10-02 Classification: LIGASE Ligands: AMP, NA, MG |
 |
Organism: Yasminevirus sp. gu-2018
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-10-02 Classification: LIGASE Ligands: ATP |
 |
Organism: Geobacillus stearothermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-08-18 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: DTP, SO4, MG |
 |
Crystal Structure Of The Cas6 Domain Of Marinomonas Mediterranea Mmb-1 Cas6-Rt-Cas1 Fusion Protein
Organism: Escherichia coli o157:h7, Marinomonas mediterranea mmb-1
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2018-10-17 Classification: HYDROLASE Ligands: GOL, SO4 |
 |
Structure Of A Thermostable Group Ii Intron Reverse Transcriptase With Template-Primer And Its Functional And Evolutionary Implications (Rt/Duplex (Nat))
Organism: Geobacillus stearothermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2017-11-29 Classification: RNA binding protein/RNA/DNA Ligands: DTP, SO4, MG |
 |
Structure Of A Thermostable Group Ii Intron Reverse Transcriptase With Template-Primer And Its Functional And Evolutionary Implications (Rt/Duplex (Se-Met))
Organism: Geobacillus stearothermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2017-11-29 Classification: RNA binding protein/RNA/DNA Ligands: DTP, MG |
 |
Structure Of A Thermostable Group Ii Intron Reverse Transcriptase With Template-Primer And Its Functional And Evolutionary Implications (Duplex Only)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2017-11-29 Classification: DNA/RNA |
 |
Crystal Structure Of A C-Terminally Truncated Aspergillus Nidulans Mitochondrial Tyrosyl-Trna Synthetase
Organism: Emericella nidulans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-06 Classification: LIGASE Ligands: TYR |
 |
Crystal Structure Of A C-Terminally Truncated Coccidioides Posadasii Mitochondrial Tyrosyl-Trna Synthetase
Organism: Coccidioides posadasii (strain c735)
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2016-04-06 Classification: LIGASE Ligands: TYR |
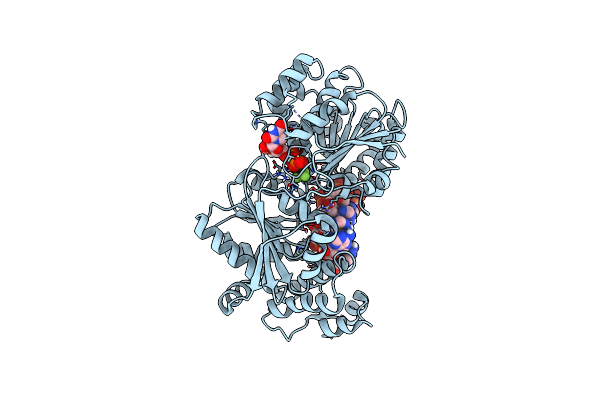 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2015-01-28 Classification: RNA BINDING PROTEIN/RNA Ligands: BEF, UDP, MG |
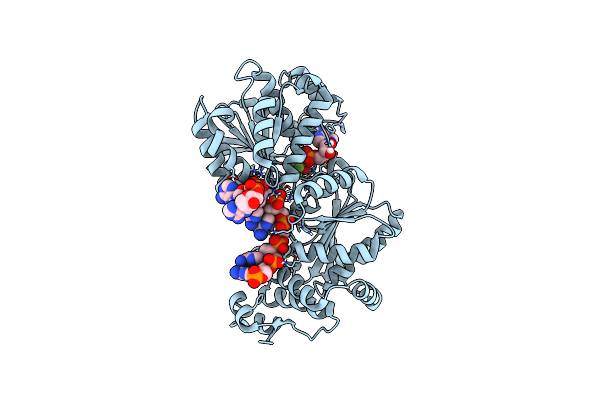 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2014-12-31 Classification: HYDROLASE/DNA Ligands: MG, BEF, ADP |
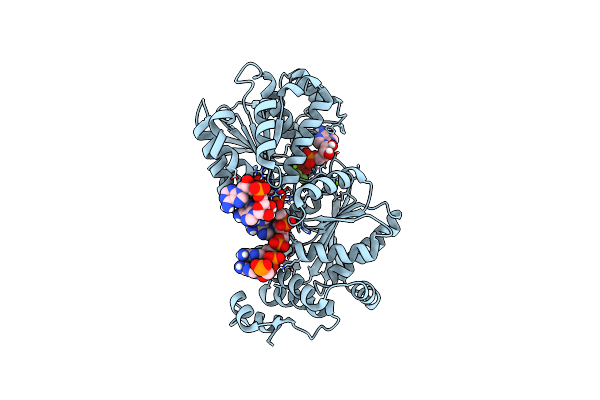 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-12-31 Classification: RNA BINDING PROTEIN/RNA Ligands: MG, BEF, ADP |
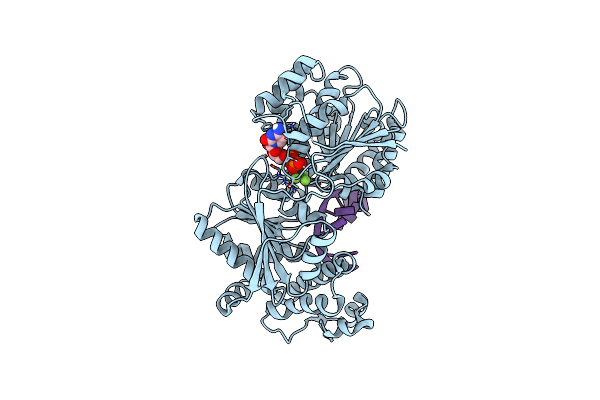 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2014-12-31 Classification: RNA BINDING PROTEIN/RNA Ligands: BEF, CDP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-12-31 Classification: RNA BINDING PROTEIN/RNA Ligands: GDP, BEF, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2012-08-29 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2012-08-29 Classification: RNA-BINDING PROTEIN/DNA,RNA |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2011-10-12 Classification: HYDROLASE/RNA Ligands: ANP, MG |
 |
Structure Of Mss116P (Nte And C-Tail Double Deletion) Bound To Ssrna And Amp-Pnp
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2011-10-12 Classification: HYDROLASE/RNA Ligands: ANP, MG |

