Search Count: 19
 |
Organism: Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2025-03-19 Classification: STRUCTURAL PROTEIN |
 |
Organism: Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-03-19 Classification: STRUCTURAL PROTEIN |
 |
Organism: Helicobacter pylori cpy1124, Helicobacter pylori (strain atcc 700392 / 26695), Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2020-05-13 Classification: CHAPERONE |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-13 Classification: MOTOR PROTEIN |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-09-13 Classification: TRANSFERASE/MOTOR PROTEIN Ligands: FLC |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2013-05-08 Classification: MOTOR PROTEIN |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-05-08 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of A Putative Ntp Pyrophosphohydrolase (Exig_1061) From Exiguobacterium Sp. 255-15 At 1.78 A Resolution
Organism: Exiguobacterium sibiricum 255-15
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2010-07-21 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-06-30 Classification: CHAPERONE |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2010-06-30 Classification: CHAPERONE |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2010-06-30 Classification: CHAPERONE |
 |
Crystal Structure Of Putative Sufu (Suppressor Of Fused Protein) Homolog (Yp_208451.1) From Neisseria Gonorrhoeae Fa 1090 At 1.40 A Resolution
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-01-26 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Crystal Structure Of Putative Cell Invasion Protein With Mac/Perforin Domain (Np_812351.1) From Bacteriodes Thetaiotaomicron Vpi-5482 At 2.46 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2009-11-24 Classification: CELL INVASION Ligands: CL, EDO, MPD |
 |
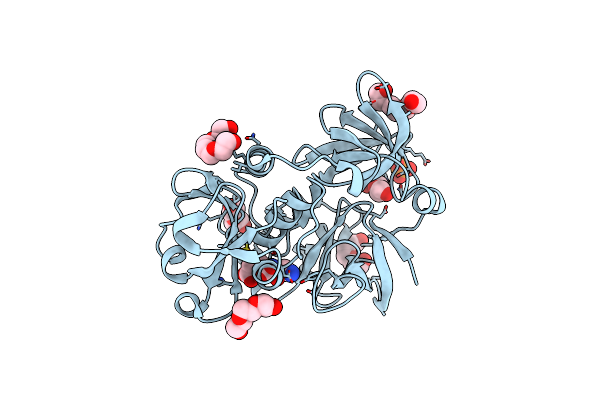
Crystal Structure Of A Putative Glycoside Hydrolase (Bt_2081) From Bacteroides Thetaiotaomicron Vpi-5482 At 2.05 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-05-26 Classification: Ca-BINDING PROTEIN Ligands: CA, NA, CAC, ACT, PEG, P6G |
 |
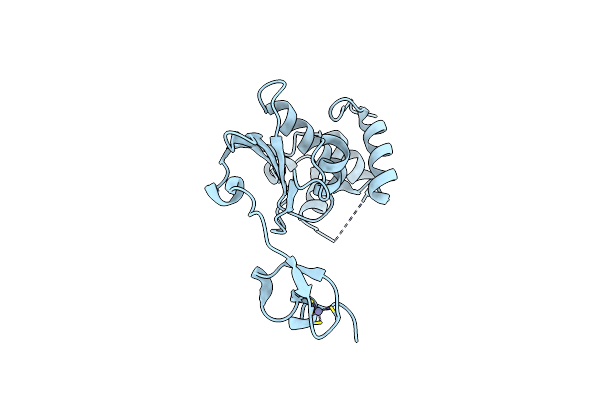
Crystal Structure Of A Nlpc/P60 Family Protein (Bce_2878) From Bacillus Cereus Atcc 10987 At 1.79 A Resolution
Organism: Bacillus cereus atcc 10987
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2009-05-05 Classification: HYDROLASE Ligands: ALA, DGL, PO4, PG4 |
 |
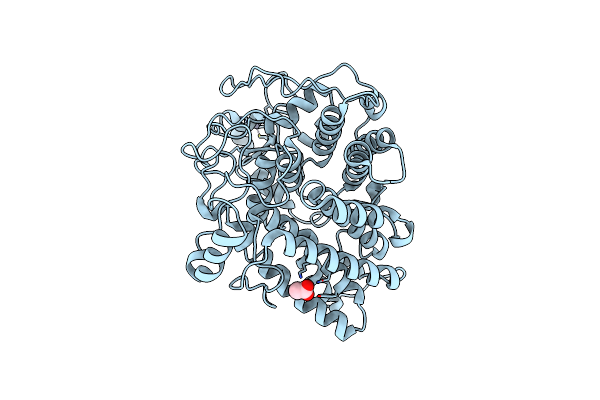
Crystal Structure Of A Tungsten Formylmethanofuran Dehydrogenase Subunit E (Fmde)-Like Protein (Syn_00638) From Syntrophus Aciditrophicus At 1.90 A Resolution
Organism: Syntrophus aciditrophicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-05-20 Classification: METAL BINDING PROTEIN Ligands: ZN, CL |
 |
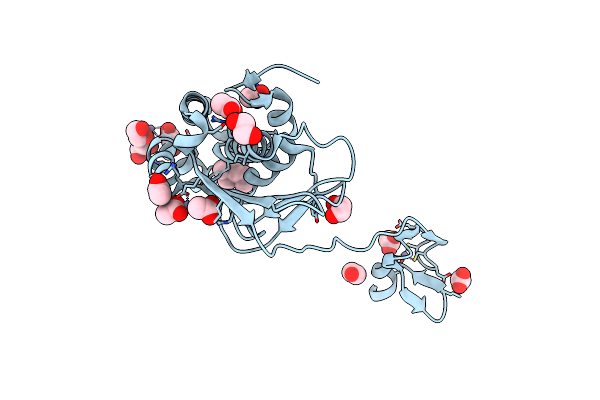
Crystal Structure Of A Susd Homolog (Bt_3984) From Bacteroides Thetaiotaomicron Vpi-5482 At 1.70 A Resolution
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-03-18 Classification: METAL BINDING PROTEIN Ligands: ZN, ACT |
 |
Crystal Structure Of A Putative Formylmethanofuran Dehydrogenase Subunit E (Ta1109) From Thermoplasma Acidophilum At 1.87 A Resolution
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2006-06-13 Classification: OXIDOREDUCTASE Ligands: ZN, UNL, EDO, ACY |
 |
Crystal Structure Of A Formylmethanofuran Dehydrogenase Subunit E-Like Protein (Dhaf_2992) From Desulfitobacterium Hafniense Dcb-2 At 1.45 A Resolution
Organism: Desulfitobacterium hafniense
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-04-18 Classification: METAL BINDING PROTEIN Ligands: ZN, NI, EDO |

