Search Count: 218
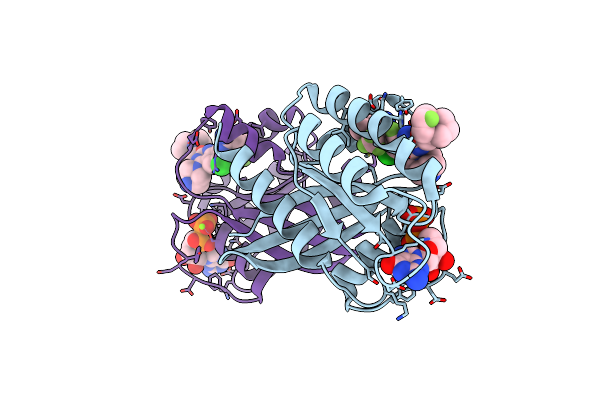 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-11-08 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, VM9 |
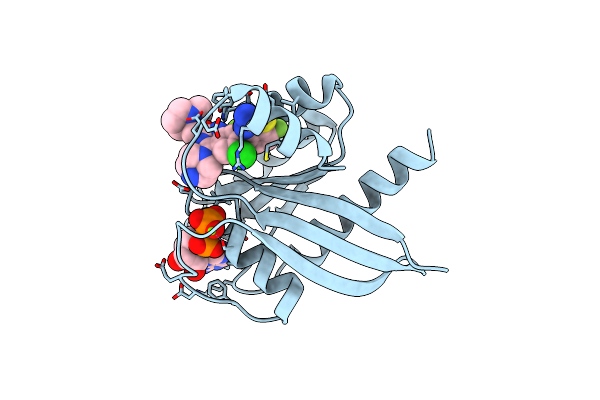 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-11-08 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, VQT |
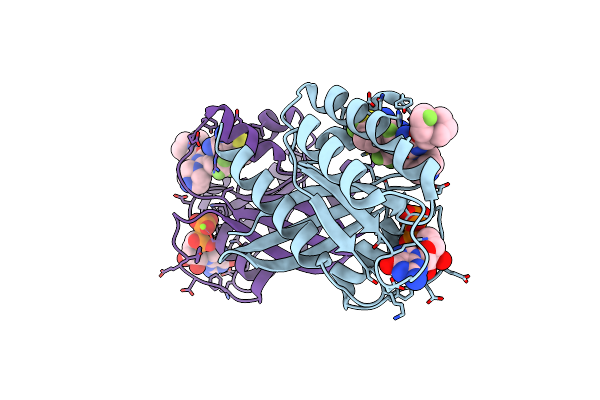 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2023-11-08 Classification: ONCOPROTEIN/INHIBITOR Ligands: GDP, MG, VR5 |
 |
Organism: Rana temporaria
Method: SOLUTION NMR Release Date: 2023-09-06 Classification: ANTIMICROBIAL PROTEIN |
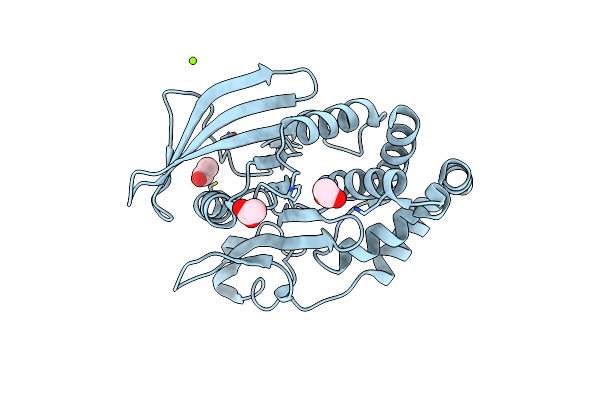 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-01-19 Classification: HYDROLASE Ligands: ACT, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-01-19 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: WOV, MG |
 |
Crystal Structure Of Pi3-E12 Fab, An Antibody Against Human Parainfluenza Virus Type Iii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2021-05-05 Classification: IMMUNE SYSTEM Ligands: PEG, EDO, SO4, NH4 |
 |
Unusual Posttranslational Modifications Revealed In Crystal Structures Of Diatom Rubisco.
Organism: Chaetoceros socialis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-06-27 Classification: LYASE Ligands: MG, CAP, EDO |
 |
Organism: Skeletonema marinoi
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-06-27 Classification: LYASE Ligands: MG, CAP, EDO |
 |
Organism: Thalassiosira hyalina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-03-14 Classification: PHOTOSYNTHESIS Ligands: MG, CAP, EDO |
 |
Organism: Thalassiosira antarctica var. borealis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-02-14 Classification: PHOTOSYNTHESIS Ligands: MG, CAP, EDO |
 |
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: PO4, EDO |
 |
1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase From Vibrio Vulnificus In Complex With Mn(2+)
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: CL, EDO, MN |
 |
1-Deoxy-D-Xylulose 5-Phosphate Reductoisomerase From Vibrio Vulnificus In Complex Arginine
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: EDO, ARG, CL, SRT |
 |
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: EDO |
 |
Organism: Vibrio vulnificus (strain cmcp6)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-07-12 Classification: OXIDOREDUCTASE Ligands: EDO, PO4, MN |
 |
Organism: Staphylococcus aureus subsp. aureus cn1
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2017-01-18 Classification: METAL BINDING PROTEIN Ligands: ZN, ACT |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-05-06 Classification: UNKNOWN FUNCTION Ligands: CL |
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-05-06 Classification: UNKNOWN FUNCTION |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2015-05-06 Classification: TRANSFERASE Ligands: ZN, PO4, SCN |

