Search Count: 49
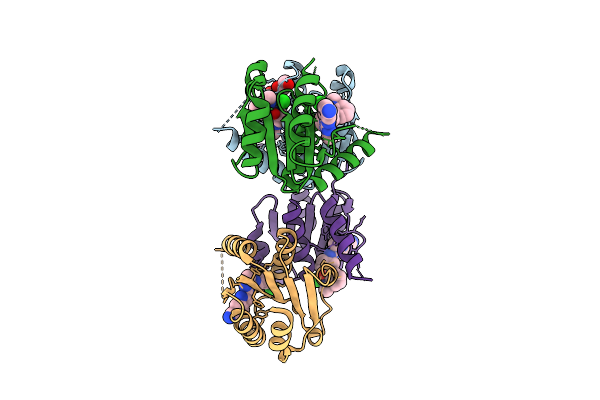 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: EOH, EDO, A1I1M, CL |
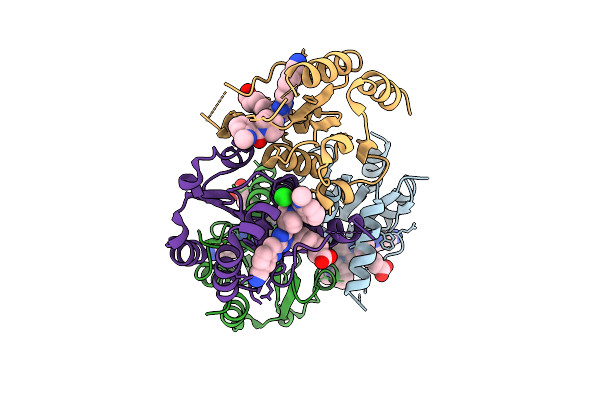 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: CL, A1I1U |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1JFX |
 |
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2024-09-04 Classification: DNA BINDING PROTEIN Ligands: SO4 |
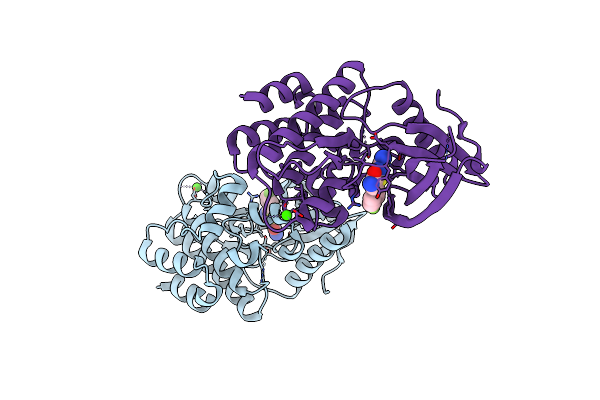 |
Human Tyk2 Pseudokinase Domain (575-869) In Complex With 5-(4-Fluoro-Phenyl)-2-Ureido-Thiophene-3-Carboxylic Acid Amide.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2021-04-14 Classification: SIGNALING PROTEIN Ligands: NM7, CA |
 |
Organism: Pseudopleuronectes americanus
Method: SOLUTION NMR Release Date: 2020-12-09 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Pseudopleuronectes americanus
Method: SOLUTION NMR Release Date: 2020-12-09 Classification: ANTIMICROBIAL PROTEIN |
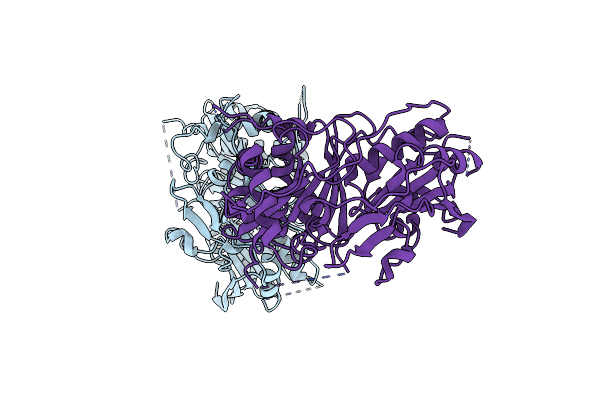 |
Organism: Selaginella moellendorffii
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2018-10-03 Classification: TRANSFERASE |
 |
X-Ray Structure Of The Complex Between Human Alpha-Thrombin And Modified 15-Mer Dna Aptamer Containing 5-(3-(2-(1H-Indol-3-Yl)Acetamide-N-Yl)-1-Propen-1-Yl)-2'-Deoxyuridine Residue
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2017-10-25 Classification: Hydrolase/DNA Ligands: K, 0G6, NA, NAG |
 |
X-Ray Structure Of The Complex Between Human Alpha-Thrombin And Modified 15-Mer Dna Aptamer Containing 5-(3-(Acetamide-N-Yl)-1-Propen-1-Yl)-2'-Deoxyuridine Residue
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2017-10-18 Classification: HYDROLASE/DNA Ligands: K, 0G6, NA, NAG |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Methyl-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, 8RK, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-2-Oxo-6-[(1R)-1-(Pyridin-2-Yl)Ethoxy]-2,3-Dihydro-1,3-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8R8 |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-6-[(1R)-1-(Pyridin-2-Yl)Ethoxy]-1,2-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8R5 |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-{5-Chloro-6-[(1R)-1-(6-Methylpyridazin-3-Yl)Ethoxy]-1,2-Benzoxazol-3-Yl}Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, 8RB |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With The Enzyme Substrate L-Kynurenine
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-06-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, GOL, KYN |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5,6-Dichloro-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2017-06-14 Classification: OXIDOREDUCTASE Ligands: FAD, CL, JHY, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Ethoxy-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: FAD, CL, 8EQ, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-(5-Chloro-6-Cyclopropoxy-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl)Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: FAD, CL, FYK, GOL |
 |
Pseudomonas Fluorescens Kynurenine 3-Monooxygenase (Kmo) In Complex With 3-[5-Chloro-6-(Cyclobutylmethoxy)-2-Oxo-2,3-Dihydro-1,3-Benzoxazol-3-Yl]Propanoic Acid
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-04-19 Classification: OXIDOREDUCTASE Ligands: FAD, CL, OK1, GOL |

