Search Count: 21
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, A1BM0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, PGE, A1BMX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, PGE, EDO, A1BMY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: ZN, PD9 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: 7YC, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-03-21 Classification: nuclear protein/antagonist Ligands: F3D, EDO, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2018-03-21 Classification: nuclear protein/antagonist Ligands: F3D, EDO |
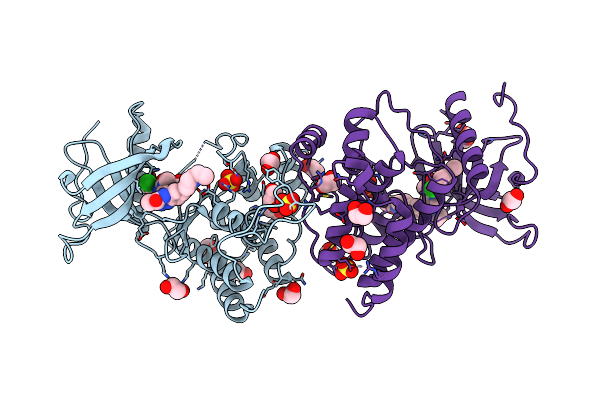 |
Crystal Structure Of Fgfr1-Y563C (Fgfr4 Surrogate) Covalently Bound To H3B-6527
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-05-24 Classification: transferase/transferase inhibitor Ligands: 9ES, SO4, EDO |
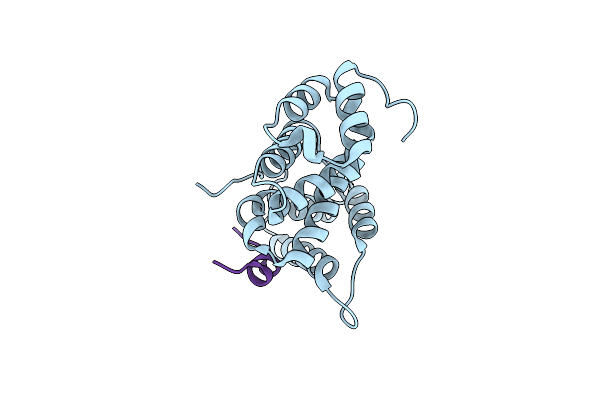 |
Crystal Structure Of Human Cnot1 Mif4G Domain In Complex With A Ttp Peptide
Organism: Homo sapiens, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-05-08 Classification: PROTEIN BINDING |
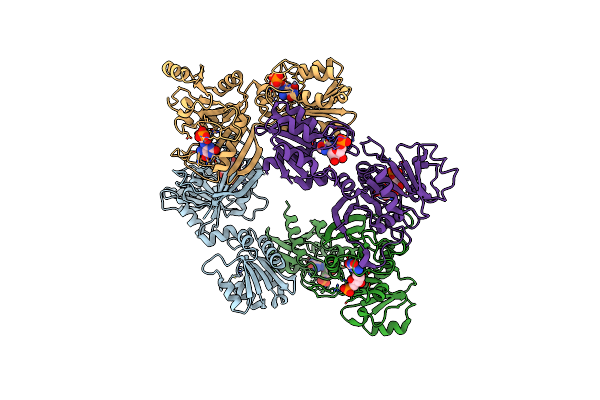 |
Complex Structure Of Bacillus Subtilis Ribg: The Deamination Process In Riboflavin Biosynthesis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2013-02-20 Classification: HYDROLASE, OXIDOREDUCTASE Ligands: ZN, AI9, AOF |
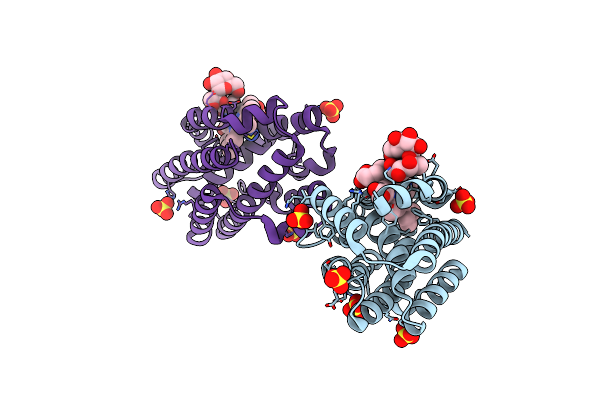 |
Crystal Structure Of The Hmuo, Heme Oxygenase From Corynebacterium Diphtheriae, In Complex With Azide-Bound Verdoheme
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2011-03-09 Classification: OXIDOREDUCTASE Ligands: AZI, SO4, VEA |
 |
Functional Roles Of The 6-S-Cysteinyl, 8 Alpha-N1-Histidyl Fad In Glucooligosaccharide Oxidase From Acremonium Strictum
Organism: Acremonium strictum
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2009-06-30 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, ZN |
 |
The Crystal Structure Of An Acremonium Strictum Glucooligosaccharide Oxidase Reveals A Novel Flavinylation
Organism: Acremonium strictum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE Ligands: NAG, ZN, FAD |
 |
Crystal Structure Of Glucooligosaccharide Oxidase From Acremonium Strictum: A Novel Flavinylation Of 6-S-Cysteinyl, 8Alpha-N1-Histidyl Fad
Organism: Acremonium strictum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE Ligands: NAG, ZN, FAD, ABL |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: ACT, ZN, CU |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: ACT, ZN |
 |
The Crystal Structure Of The D-Aminoacylase D366A Mutant In Complex With 100Mm Zncl2
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: ACT, ZN |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: ACT, ZN, CU |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: ACT, ZN, CD |
 |
The Functional Role Of The Binuclear Metal Center In D-Aminoacylase. One-Metal Activation And Second-Metal Attenuation
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2004-04-20 Classification: HYDROLASE Ligands: ACT, ZN |

