Search Count: 16
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-01-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, MG, A1BEI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, A1BEJ, MG |
 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-05-25 Classification: OXIDOREDUCTASE Ligands: NAI, DMS, MES |
 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-05-25 Classification: OXIDOREDUCTASE Ligands: NAI, DMS |
 |
Organism: Yersinia pestis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-05-25 Classification: OXIDOREDUCTASE Ligands: NAI, DMS |
 |
The Effect Of Isoleucine To Alanine Mutation On Inha Enzyme Crystallization Pattern And Inhibition By Ligand Pt70 (Tcu)
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-08-19 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Compensation Of The Effect Of Isoleucine To Alanine Mutation By Designed Inhibition In The Inha Enzyme
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:3.41 Å Release Date: 2015-08-12 Classification: OXIDOREDUCTASE Ligands: NAD, 53K |
 |
The Effect Of Valine To Alanine Mutation On Inha Enzyme Crystallization Pattern And Substrate Binding Loop Conformation And Flexibility
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE Ligands: NAD, TCU, NA |
 |
The Effect Of Isoleucine To Alanine Mutation On Inha Enzyme Crystallization Pattern And Substrate Binding Loop Conformation And Flexibility
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-08-05 Classification: OXIDOREDUCTASE Ligands: NAD, TCU, ETE, EPE |
 |
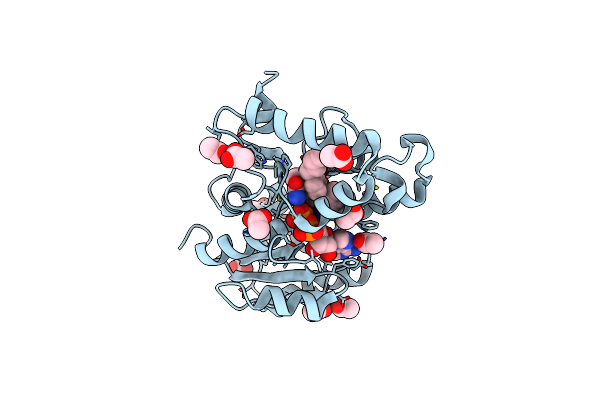
Crystal Structure Of Mycobacterium Tuberculosis Inha In Complex With Inhibitor Pt92
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 2TK |
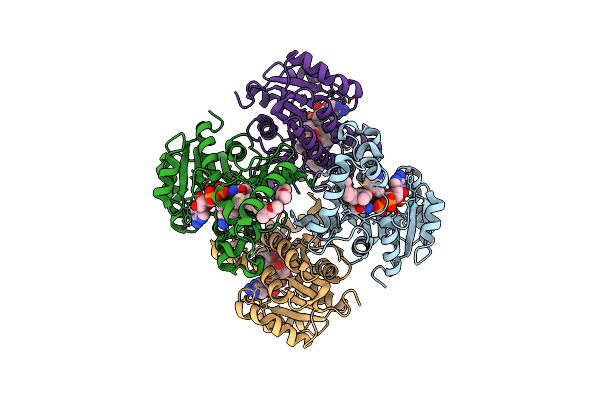 |
Multiple Binding Modes Of Inhibitor Pt155 To The Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha Within A Tetramer
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2014-04-30 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: NAD, 1S5, 2NV |
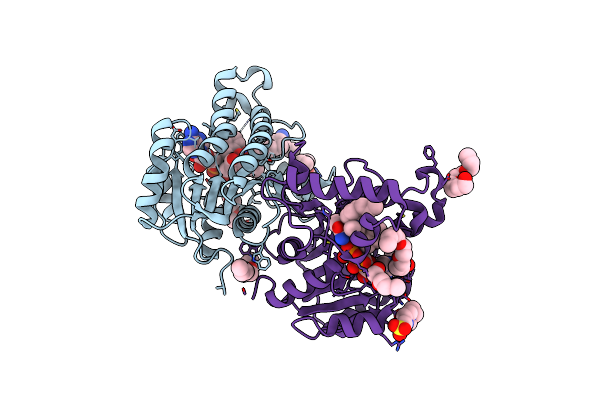 |
Substrate-Like Binding Mode Of Inhibitor Pt155 To The Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 1S5, CL, 2NV, EPE |
 |
Substrate-Binding Loop Movement With Inhibitor Pt10 In The Tetrameric Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 1TN |
 |
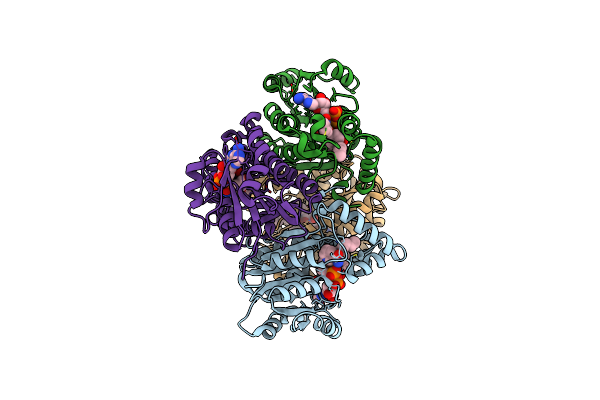
Competition Of The Small Inhibitor Pt91 With Large Fatty Acyl Substrate Of The Mycobacterium Tuberculosis Enoyl-Acp Reductase Inha By Induced Substrate-Binding Loop Refolding
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-04-30 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, 1US |
 |
Crystal Structure Of Mycobacterium Tuberculosis Inha In Complex With Inhibitor Pt119 In 2.4 M Acetate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-04-23 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, JUS, ACT |
 |
Crystal Structure Of Bacillus Subtilis Guanine Deaminase. The First Domain-Swapped Structure In The Cytidine Deaminase Superfamily
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2004-07-13 Classification: HYDROLASE Ligands: ZN, IMD |

