Search Count: 26
 |
Structure Of A Complex Between Pasteurella Multocida Surface Lipoprotein, Pmslp-1, And Bovine Complement Factor I
Organism: Pasteurella multocida 36950, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: NAG |
 |
Crystal Structure Of A Slam-Dependent Surface Lipoprotein, Pmslp, In Pasteurella Multocida
Organism: Pasteurella multocida 36950
Method: X-RAY DIFFRACTION Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: SE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-04-03 Classification: METAL TRANSPORT Ligands: ATP, TL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-04-03 Classification: METAL BINDING PROTEIN Ligands: ATP, NA |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ADP, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Mgcl2
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Edta And Egta
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Edta And Egta, Vertical C2 Symmetry Axis
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Edta And Egta, C1 Symmetry
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-01-24 Classification: METAL BINDING PROTEIN Ligands: SO4, ZN |
 |
Organism: Araneus ventricosus
Method: SOLUTION NMR Release Date: 2022-04-06 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of A Class I Preq1 Riboswitch Aptamer (Ab13-14) Complexed With A Cognate Ligand-Derived Photoaffinity Probe
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2021-11-17 Classification: RNA Ligands: J0C, SO4, MG |
 |
Crystal Structure Of A Class I Preq1 Riboswitch Aptamer (Wild-Type) Complexed With A Cognate Ligand-Derived Photoaffinity Probe
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-11-17 Classification: RNA Ligands: J0C, SO4 |
 |
Organism: Actinobacillus pleuropneumoniae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-08-25 Classification: TRANSPORT PROTEIN Ligands: PLP |
 |
N-Terminal Domain (Ntd) Solution Structure Of Aciniform Spidroin (Acspn) From Nephila Antipodiana.
Organism: Nephila antipodiana
Method: SOLUTION NMR Release Date: 2020-07-22 Classification: STRUCTURAL PROTEIN |
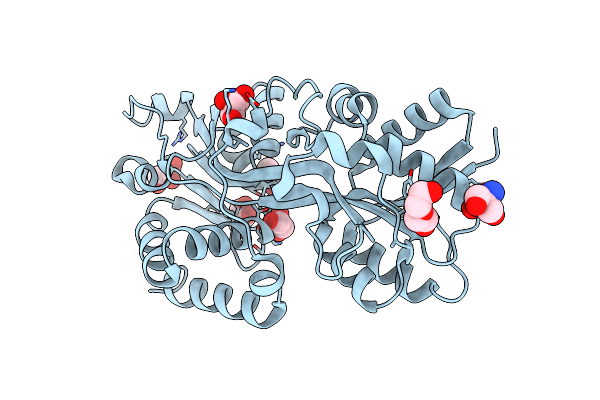 |
Structure Of The Periplasmic Binding Protein Afua From Actinobacillus Pleuropneumoniae (Apo Form)
Organism: Actinobacillus pleuropneumoniae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-08-12 Classification: TRANSPORT PROTEIN Ligands: GOL, PEG, TRS, CL |
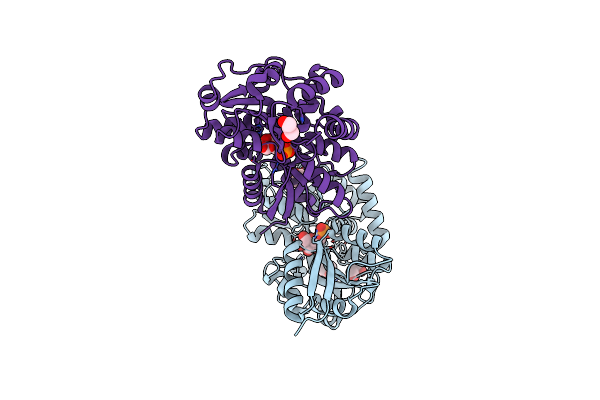 |
Structure Of The Periplasmic Binding Protein Afua From Actinobacillus Pleuropneumoniae (Endogenous Glucose-6-Phosphate And Mannose-6-Phosphate Bound)
Organism: Actinobacillus pleuropneumoniae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-08-12 Classification: TRANSPORT PROTEIN Ligands: G6P, M6P, GOL, CL |
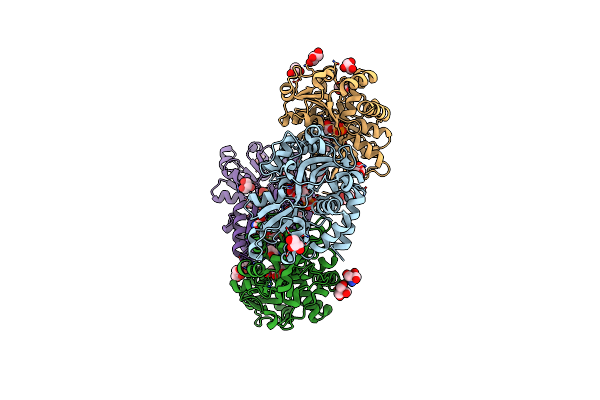 |
Structure Of The Periplasmic Binding Protein Afua From Actinobacillus Pleuropneumoniae (Exogenous Fructose-6-Phosphate Bound)
Organism: Actinobacillus pleuropneumoniae
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2015-08-12 Classification: TRANSPORT PROTEIN Ligands: F6P, GOL, TRS, MG |
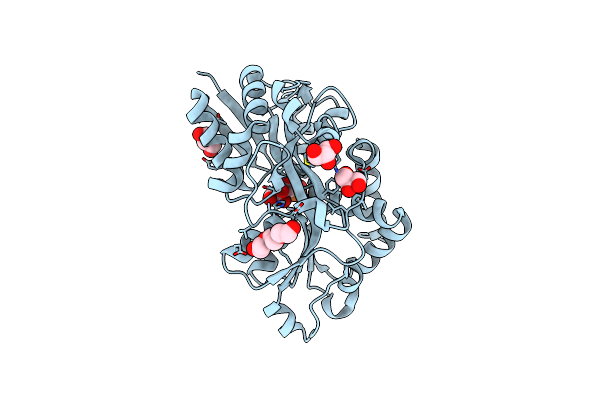 |
Structure Of The Periplasmic Binding Protein Afua From Actinobacillus Pleuropneumoniae (Exogenous Sedoheptulose-7-Phosphate Bound)
Organism: Actinobacillus pleuropneumoniae
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2015-08-12 Classification: TRANSPORT PROTEIN Ligands: S7P, GOL, PGE |
 |
Plk-1 Polo-Box Domain In Complex With Bioactive Imidazolium-Containing Phosphopeptide Macrocycle 3B
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-07-29 Classification: TRANSFERASE/TRANSFERASE INHIBITOR |

