Search Count: 35
All
Selected
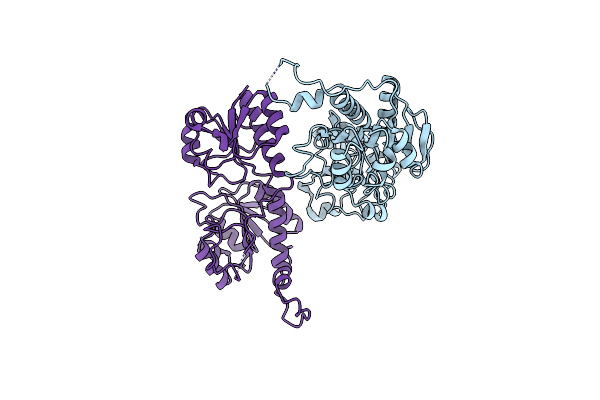 |
Crystal Structure Of Phenylpyruvate Reductase From Lactobacillus Sp. Cgmcc 9967
Organism: Lactobacillus sp. cgmcc 9967
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2023-12-20 Classification: OXIDOREDUCTASE |
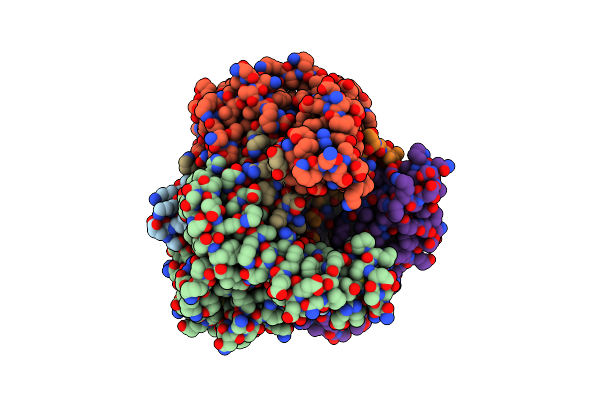 |
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2002-03-13 Classification: LYASE |
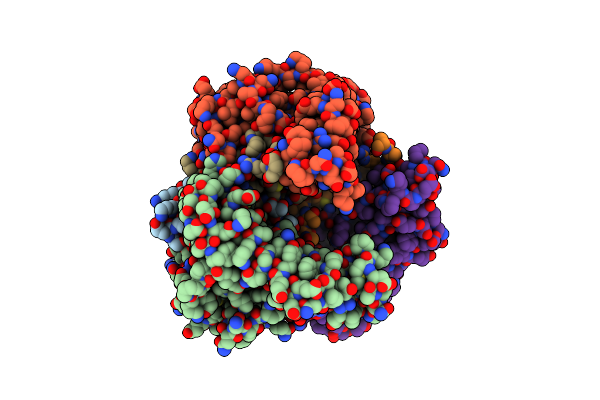 |
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2002-03-13 Classification: LYASE |
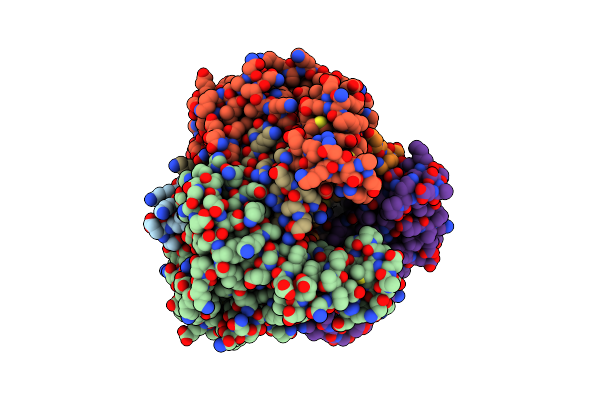 |
Structure Of The D53,54N Mutant Of Histidine Decarboxylase Bound With Histidine Methyl Ester At-170 C
Organism: Lactobacillus sp. 30a
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2002-03-13 Classification: LYASE Ligands: PVH |
 |
Structure Of The D53,54N Mutant Of Histidine Decarboxylase Bound With Histidine Methyl Ester At 25 C
Organism: Lactobacillus sp. 30a
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2002-03-13 Classification: LYASE Ligands: PVH |
 |
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2001-03-21 Classification: LYASE |
 |
Organism: Lactobacillus sp. 30a
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2000-02-26 Classification: LYASE Ligands: PLP, GTP |
 |
Crystallographic Structure Of A Plp-Dependent Ornithine Decarboxylase From Lactobacillus 30A To 3.1 Angstroms Resolution
Organism: Lactobacillus sp.
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1995-09-15 Classification: CARBOXY-LYASE Ligands: PLP |
 |
Refined Structure Of The Pyruvoyl-Dependent Histidine Decarboxylase From Lactobacillus 30A
Organism: Lactobacillus sp. 30a
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1994-01-31 Classification: CARBOXY-LYASE |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Lactobacillus sp. HMSC25A02
Method: Alphafold Release Date: Classification: NA Ligands: NA |

