Search Count: 15
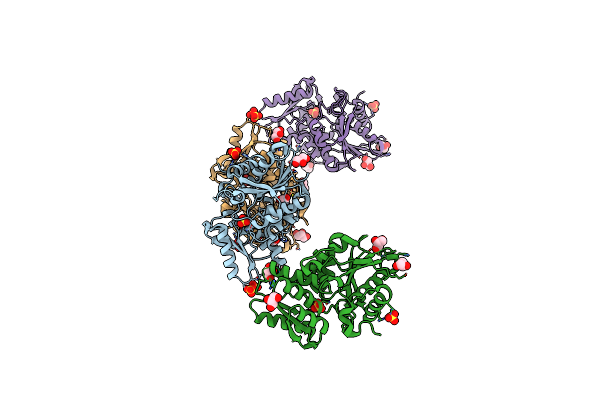 |
Crystal Structure Of Cystathionine Beta-Synthase From Lactobacillus Plantarum
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-12-07 Classification: LYASE Ligands: SO4, GOL |
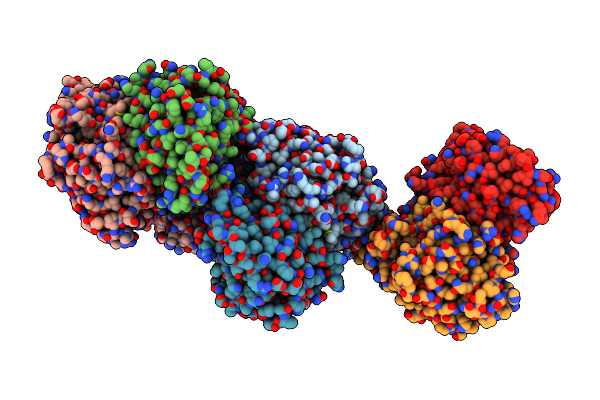 |
Crystal Structure Of K42A Mutant Of Cystathionine Beta-Synthase From Lactobacillus Plantarum In A Complex With L-Methionine
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-12-07 Classification: LYASE Ligands: AA5, SO4 |
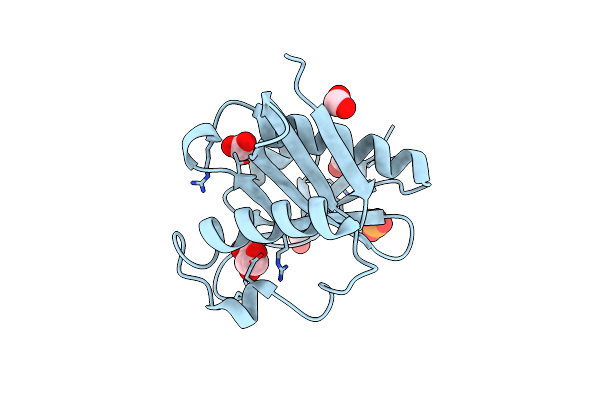 |
Crystal Structure Of Putative N-Acetyltransferase From Lactobacillus Plantarum
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2008-09-23 Classification: TRANSFERASE Ligands: FMT, GOL, PO4 |
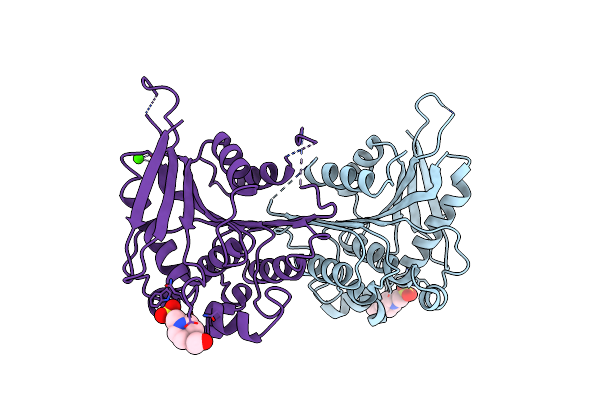 |
Crystal Structure Of Lipase/Esterase (Lp_2923) From Lactobacillus Plantarum. Northeast Structural Genomics Consortium Target Lpr108
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-06-10 Classification: HYDROLASE Ligands: EPE, CA |
 |
Crystal Structure Of The Lp_1622 Protein From Lactobacillus Plantarum. Northeast Structural Genomics Consortium Target Lpr114
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-04-15 Classification: TRANSFERASE Ligands: SO4 |
 |
Crystal Structure Of A Putative Inositol Catabolism Protein Iole (Iole, Lp_3607) From Lactobacillus Plantarum Wcfs1 At 1.85 A Resolution
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-04-08 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO, ACT |
 |
Crystal Structure Of Putative Transcription Regulator From Lactobacillus Plantarum
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-08 Classification: TRANSCRIPTION REGULATOR Ligands: GOL |
 |
Crystal Structure Of A Transcription Regulator From Lactobacillus Plantarum
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2008-04-01 Classification: TRANSCRIPTION REGULATOR Ligands: GOL |
 |
Crystal Structure Of Myo-Inositol 2-Dehydrogenase (Np_786804.1) From Lactobacillus Plantarum At 2.40 A Resolution
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-03-11 Classification: OXIDOREDUCTASE Ligands: NAD, CL, EDO |
 |
Crystal Structure Of A Putative Carboxylesterase (Lp_2923) From Lactobacillus Plantarum Wcfs1 At 1.70 A Resolution
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-01-29 Classification: HYDROLASE Ligands: EPE, PGR, PGO, CL |
 |
Crystal Structure Of A Putative Carboxylesterase (Lp_1002) From Lactobacillus Plantarum Wcfs1 At 2.09 A Resolution
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2007-12-18 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-12-04 Classification: STRUCTURAL PROTEIN |
 |
Crystal Structure Of Pyridoxamine 5'-Phosphate Oxidase-Like Protein (Np_783940.1) From Lactobacillus Plantarum At 1.55 A Resolution
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2007-11-20 Classification: OXIDOREDUCTASE Ligands: SO4, EDO |
 |
Crystal Structure Of Putative Dinucleotide-Binding Oxidoreductase (Np_786167.1) From Lactobacillus Plantarum At 1.60 A Resolution
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-10-02 Classification: OXIDOREDUCTASE Ligands: NAP, PEG |
 |
Crystal Structure Of Q88U62_Lacpl From Lactobacillus Plantarum. Northeast Structural Genomics Target Lpr71
Organism: Lactobacillus plantarum wcfs1
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-05-01 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

