Search Count: 542
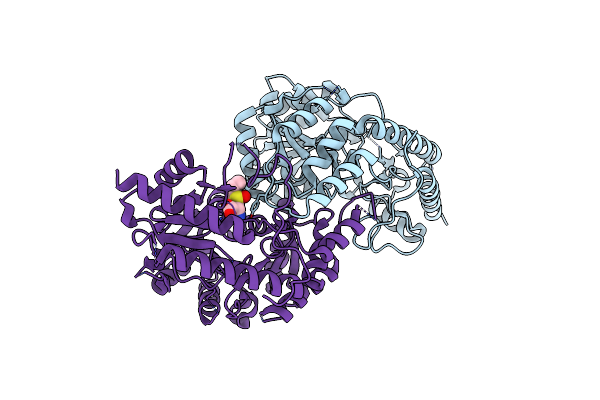 |
The Structural Complex Of Inactivated Lactobacillus Delbrueckii Cystathionine Beta-Lyase Mutant (Ldpatb A34D) With Its Substrate L-(+)-Alliin
Organism: Lactobacillus delbrueckii subsp. lactis dsm 20072
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2025-03-26 Classification: BIOSYNTHETIC PROTEIN Ligands: OO0 |
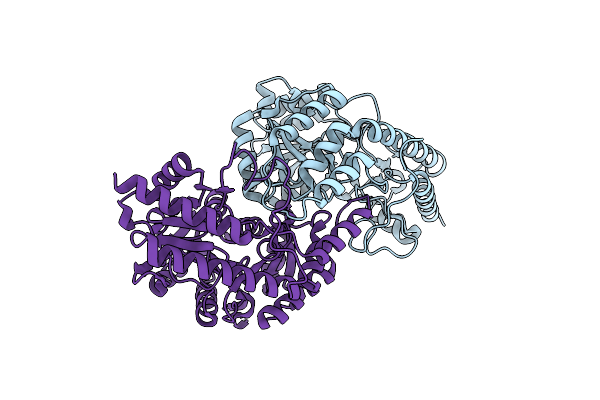 |
Three-Dimensional Structure Of Homo-Dimer Of Cystathione Beta Lyase From Lactobacillus Delbrueckii(Ldpatb)
Organism: Lactobacillus delbrueckii subsp. lactis dsm 20072
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN |
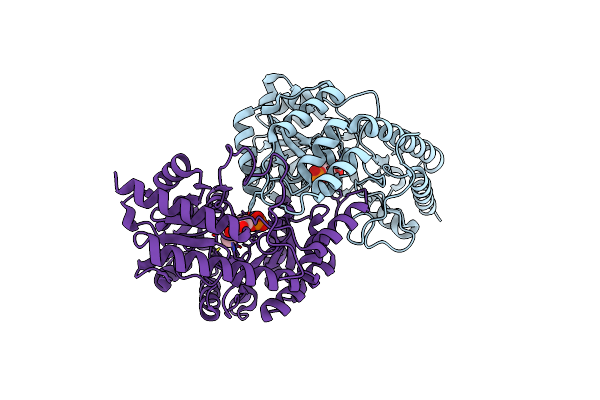 |
Three-Dimensional Structure Of Homo-Dimer Of Cystathione Beta Lyase/Plp Complex From Lactobacillus Delbrueckii(Ldpatb)
Organism: Lactobacillus delbrueckii subsp. lactis dsm 20072
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: PLP |
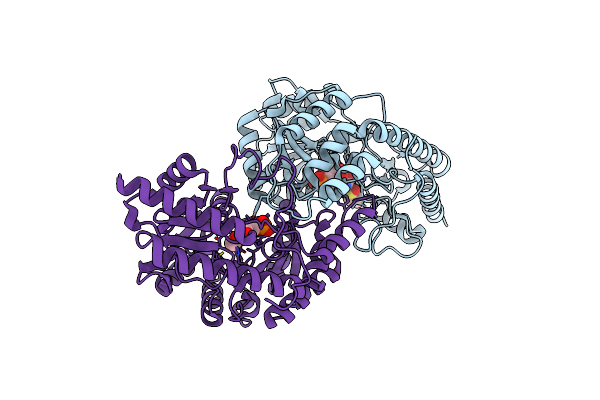 |
Three-Dimensional Structure Of Homo-Dimer Of Cystathione Beta Lyase/Plp/+L-Alliin Complex From Lactobacillus Delbrueckii(Ldpatb)
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-11-20 Classification: BIOSYNTHETIC PROTEIN Ligands: OO0, PLP |
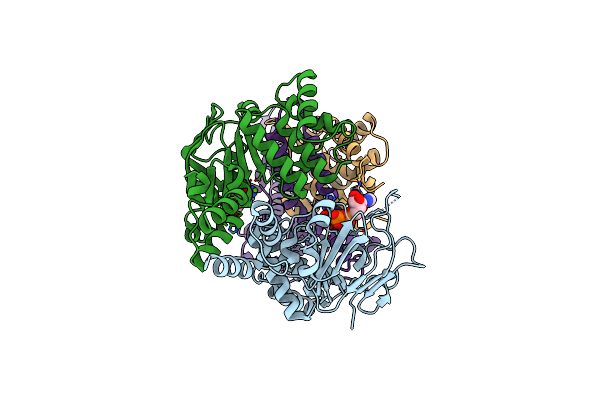 |
Cryo-Em Structure Of The Folate-Specific Ecf Transporter Complex In Msp2N2 Lipid Nanodiscs Bound To Atp And Adp
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc 11842 = jcm 1002
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: MEMBRANE PROTEIN Ligands: ATP, ADP, MG |
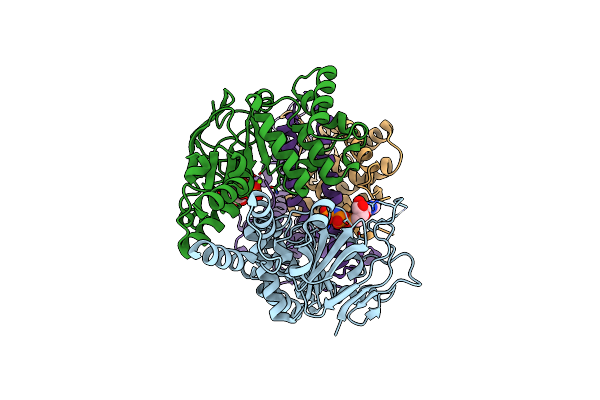 |
Cryo-Em Structure Of The Folate-Specific Ecf Transporter Complex In Msp2N2 Lipid Nanodiscs Bound To Amp-Pnp
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc 11842 = jcm 1002
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: MEMBRANE PROTEIN Ligands: ANP, MG |
 |
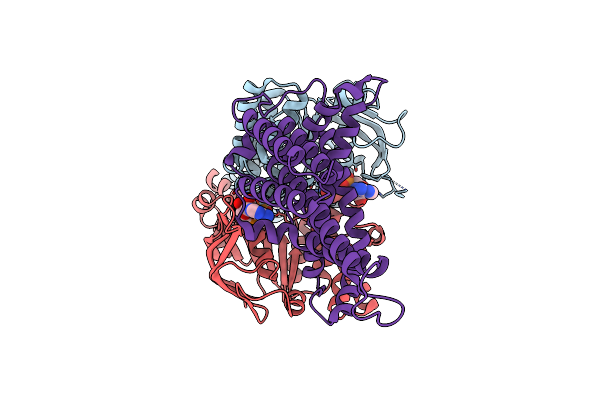
Cryo-Em Structure Of The Wild-Type Solitary Ecf Module In Msp2N2 Lipid Nanodiscs In The Atpase Open And Nucleotide-Free Conformation
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc 11842 = jcm 1002
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: MEMBRANE PROTEIN |
 |
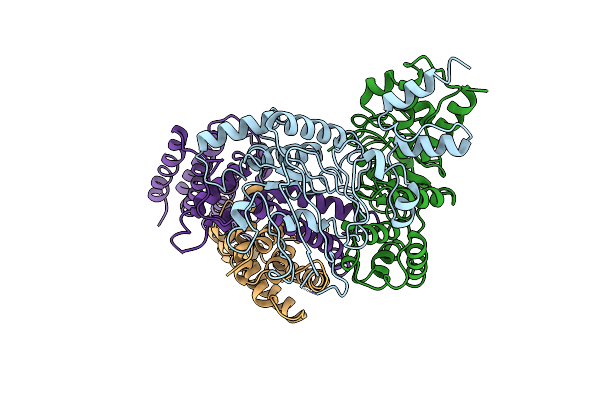
Cryo-Em Structure Of The Mutant Solitary Ecf Module 2Eq In Msp2N2 Lipid Nanodiscs In The Atpase Closed And Atp-Bound Conformation
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc 11842 = jcm 1002
Method: ELECTRON MICROSCOPY Release Date: 2023-08-02 Classification: MEMBRANE PROTEIN Ligands: ATP, MG |
 |
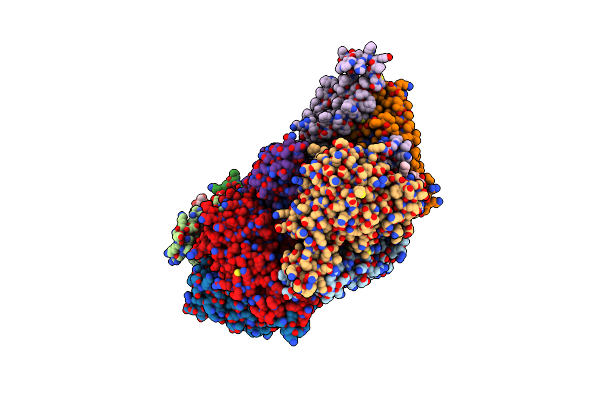
Cryo-Em Structure Of The Folate-Specific Ecf Transporter Complex In Ddm Micelles
Organism: Lactobacillus delbrueckii subsp. bulgaricus (strain atcc 11842 / dsm 20081 / jcm 1002 / nbrc 13953 / ncimb 11778)
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSPORT PROTEIN |
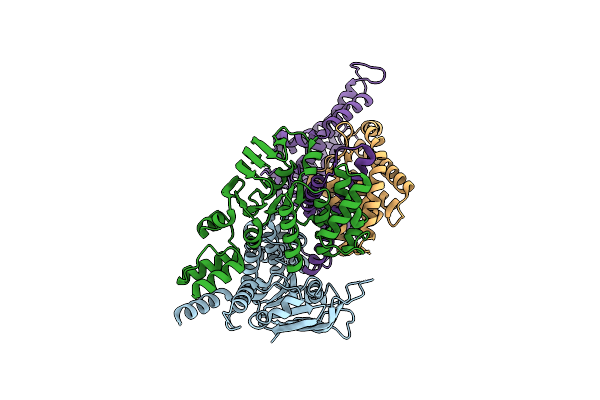 |
Cryo-Em Structure Of The Folate-Specific Ecf Transporter Complex In Msp2N2 Lipid Nanodiscs
Organism: Lactobacillus delbrueckii subsp. bulgaricus (strain atcc 11842 / dsm 20081 / jcm 1002 / nbrc 13953 / ncimb 11778)
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSPORT PROTEIN |
 |
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc 11842 = jcm 1002, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-03-03 Classification: TRANSPORT PROTEIN Ligands: PEG, LMT, CIT |
 |
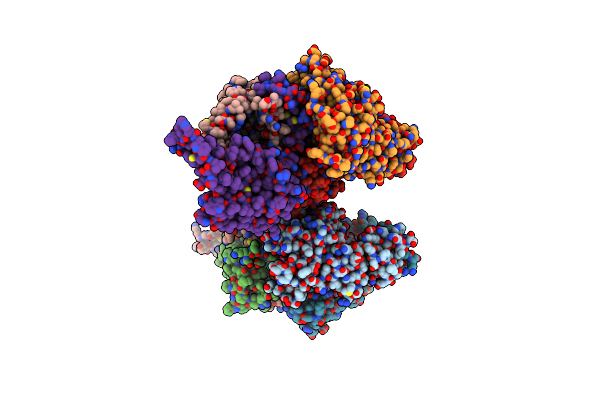
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2018-07-25 Classification: TRANSPORT PROTEIN |
 |
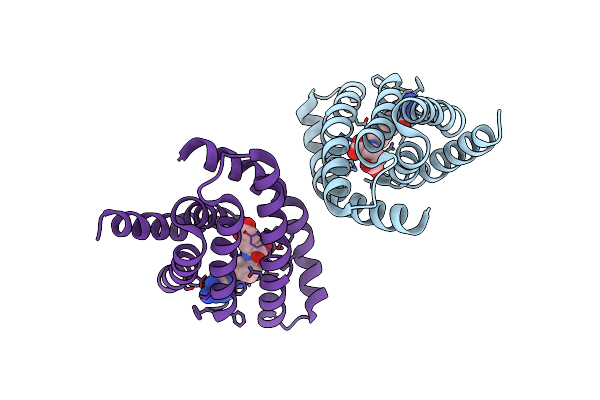
Organism: Lactobacillus delbrueckii subsp. bulgaricus (strain atcc 11842 / dsm 20081 / jcm 1002 / nbrc 13953 / ncimb 11778), Lactobacillus delbrueckii subsp. bulgaricus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2016-05-18 Classification: TRANSPORT PROTEIN |
 |
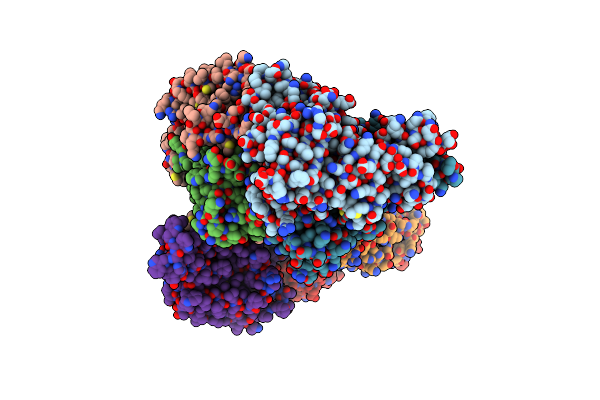
Organism: Lactobacillus delbrueckii subsp. bulgaricus
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2016-04-06 Classification: TRANSPORT PROTEIN Ligands: FOL |
 |
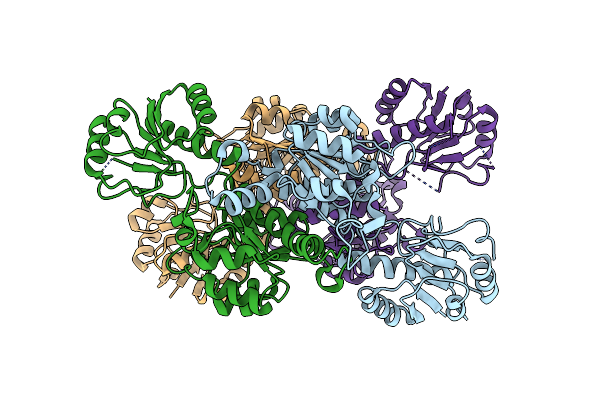
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-04-06 Classification: TRANSPORT PROTEIN Ligands: ANP |
 |
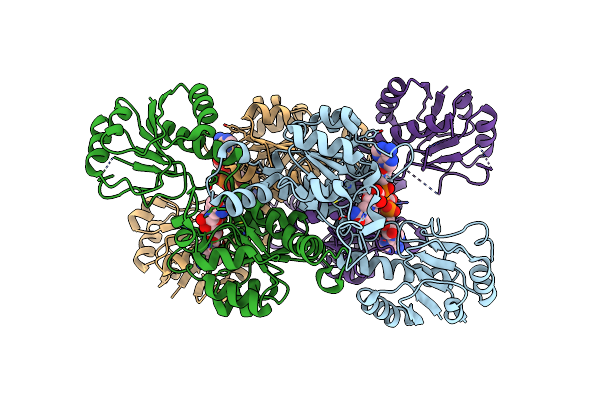
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Lactobacillus Delbrueckii Ssp. Bulgaricus
Organism: Lactobacillus delbrueckii subsp. bulgaricus
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2012-11-21 Classification: OXIDOREDUCTASE |
 |
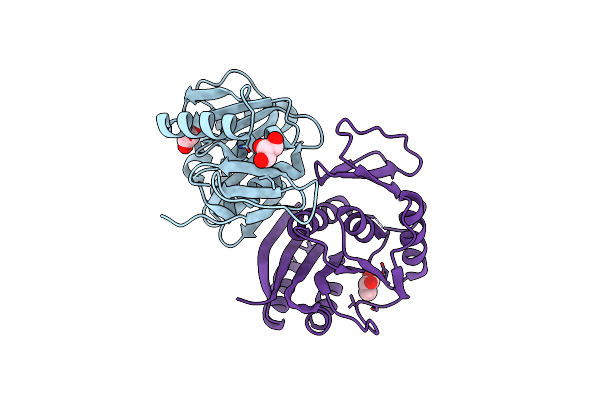
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Lactobacillus Delbrueckii Ssp. Bulgaricus: Nad Complexed Form
Organism: Lactobacillus delbrueckii subsp. bulgaricus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2012-11-21 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
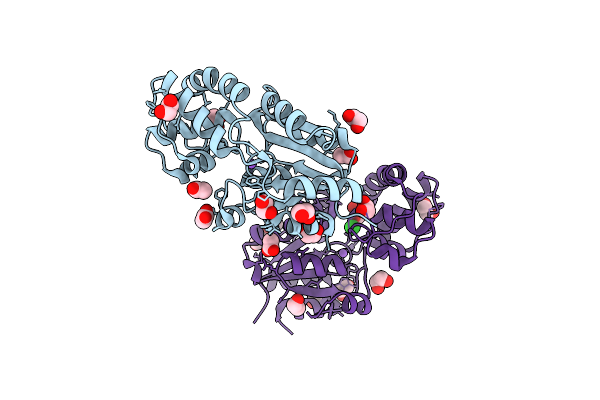
Crystal Structure Of Ribosomal Protein L11 Methylase From Lactobacillus Delbrueckii Subsp. Bulgaricus
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc baa-365
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-04-07 Classification: TRANSFERASE Ligands: GOL |
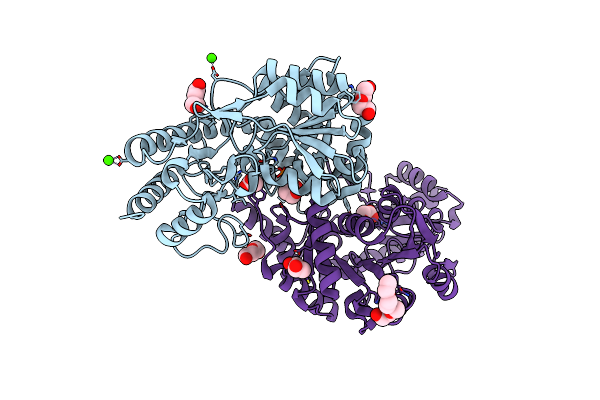 |
Crystal Structure Of A Putative Plp-Dependent Aminotransferase (Lbul_1103) From Lactobacillus Delbrueckii Subsp. At 1.61 A Resolution
Organism: Lactobacillus delbrueckii subsp. bulgaricus atcc baa-365
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: CA, CL, ACT, PEG, PG4 |
 |
Crystal Structure Of Putative Phosphoglycolate Phosphatase (Yp_619066.1) From Lactobacillus Delbrueckii Subsp. Bulgaricus Atcc Baa-365 At 1.51 A Resolution
Organism: Lactobacillus delbrueckii
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2006-08-15 Classification: HYDROLASE |

