Search Count: 51,578
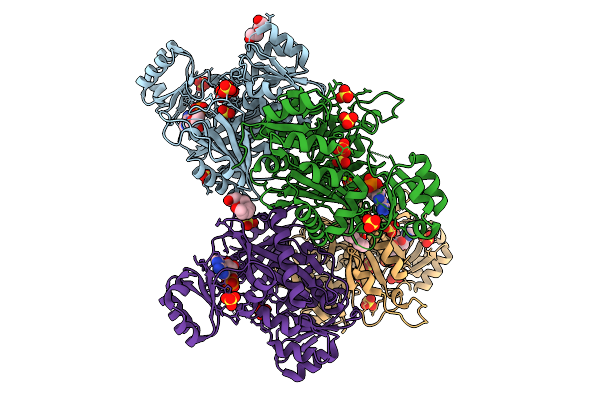 |
Crystal Structure Of Phosphoribosylaminoimidazole Carboxylase From Burkholderia Xenovorans (Amp, Adp And Sulfate Complex)
Organism: Paraburkholderia xenovorans lb400
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2026-01-21 Classification: LYASE Ligands: AMP, PEG, MG, SO4, PG4, ADP |
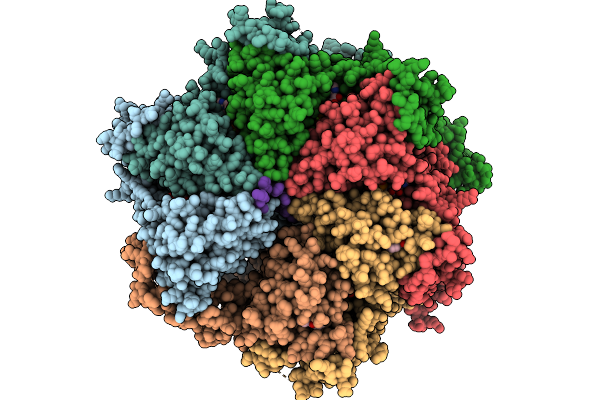 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: TRANSLOCASE Ligands: ATP, MG, A1AP0 |
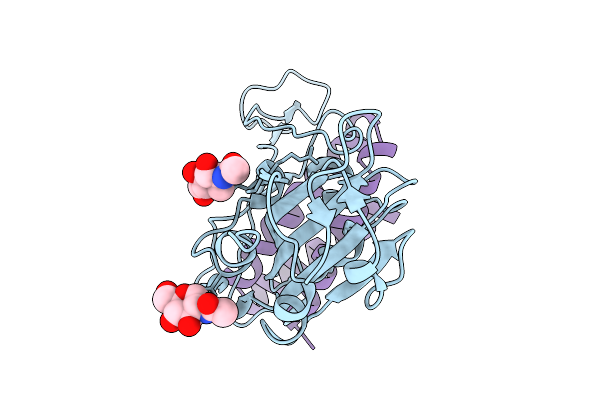 |
Local Refinement Of The Sars-Cov-2 Ba.2.86 Rbd In Complex With Tri2-2 Minibinder
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: NAG |
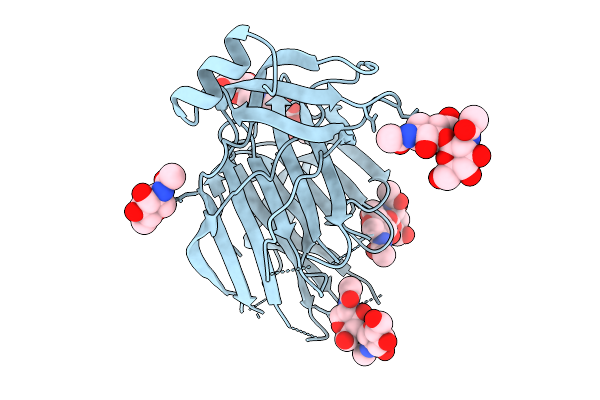 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.40 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: CL |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: CL |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: CL, NAG |
 |
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: CL, NAG |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: CL, NAG |
 |
Organism: Sterolibacterium denitrificans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-21 Classification: LYASE Ligands: PO4, MPD, IMD, GOL |
 |
Organism: Homo sapiens, Synthetic construct
Method: SOLUTION NMR Release Date: 2026-01-21 Classification: CELL CYCLE Ligands: GNP, MG |
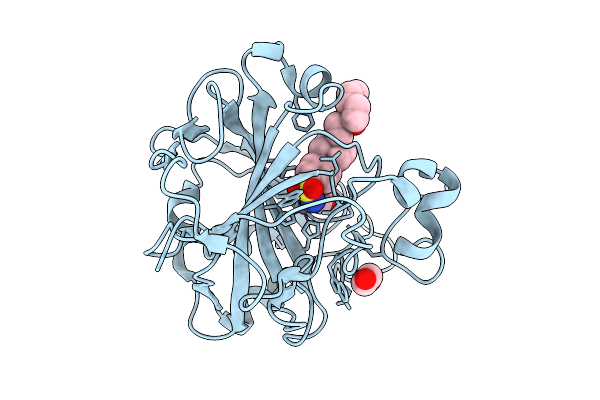 |
Crystal Structure Of Human Carbonic Anhydrase Ii In Complex With Sonepiprazole
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2026-01-21 Classification: LYASE Ligands: GOL, EDO, ZN, A1IS9 |
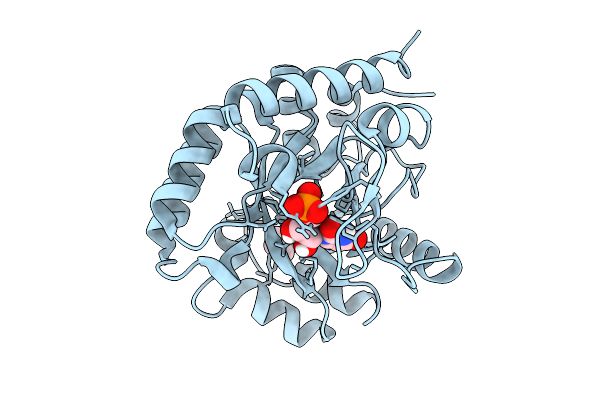 |
Variant T321A Of Orotidine 5'-Monophosphate Decarboxylase-Domain Of Human Umps In Complex With The Product Ump At 0.95 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2026-01-21 Classification: LYASE Ligands: U5P, PRO |
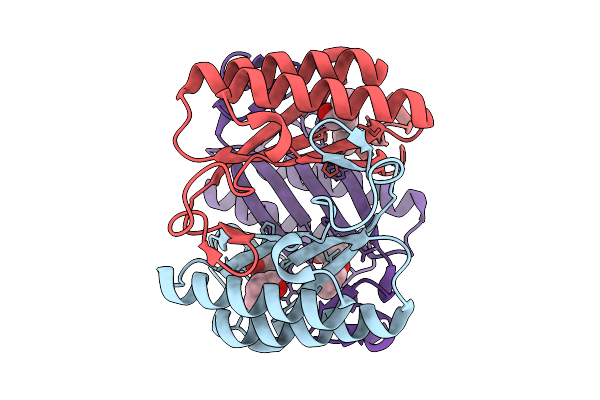 |
X-Ray Crystallographic Structure Of 4-Ot (F11) In Complex With 2-Hydroxycinnamaldehyde
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2026-01-21 Classification: LYASE Ligands: A1IZH |
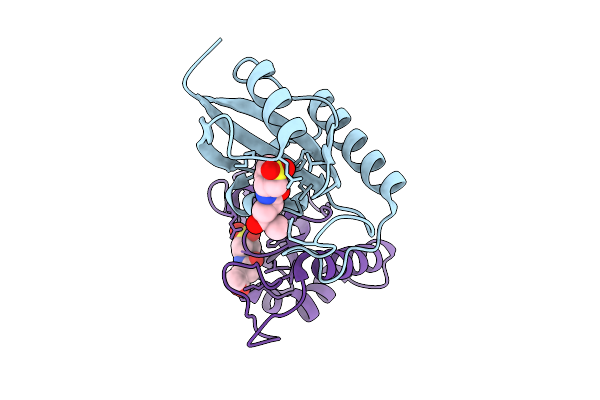 |
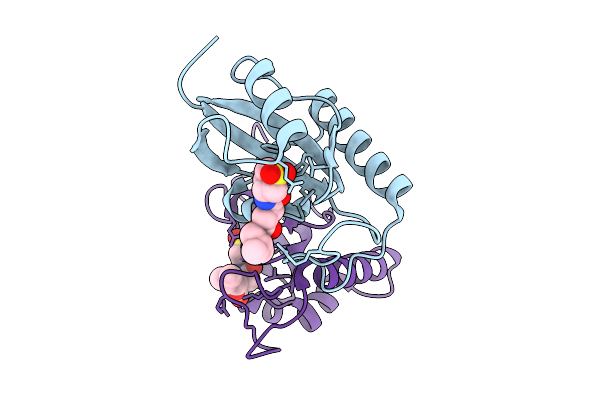
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain Complexed With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9T |
 |
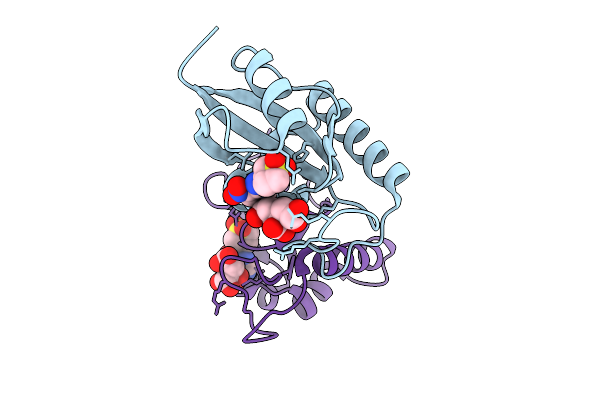
Human Pin1 (Peptidyl-Prolyl Cis-Trans Isomerase) Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9U |
 |
Crystal Structure Of Human Pin1 Catalytic Domain In Complex With A Covalent Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2026-01-21 Classification: ISOMERASE Ligands: A1D9W, CIT |
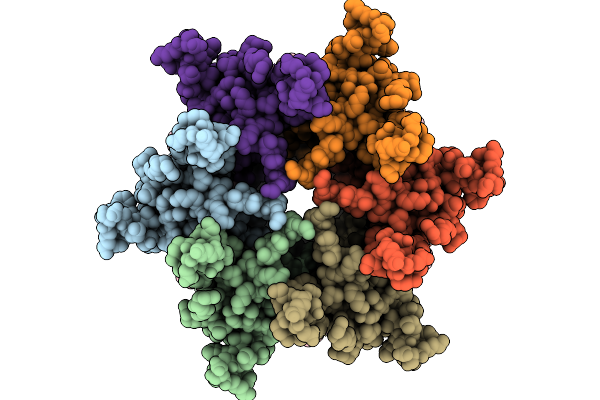 |
Hemichannel Sub-Structure Of Cx43/Gja1 Gap Junction Intercellular Channel, Treated With A 5-Molar Excess Of Carbenoxolone
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: C14 |
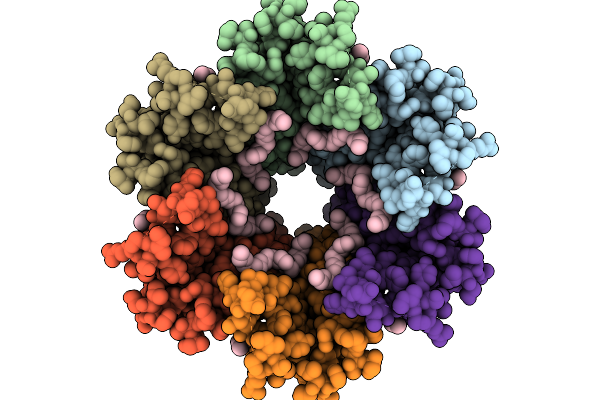 |
Hemichannel Sub-Structure Of Cx36/Gjd2 Gap Junction Intercellular Channel (Fn Conformation) In Brain Polar Lipid Nanodiscs, Treated With A 14-Fold Molar Excess Of Carbenoxolone
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: C14 |

