Search Count: 70,384
 |
Organism: Human immunodeficiency virus type 1 (new york-5 isolate)
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN Ligands: A1MCX, 1PE, SO4 |
 |
Organism: Faecalibacterium duncaniae
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: TRS, GOL, EDO, PEG |
 |
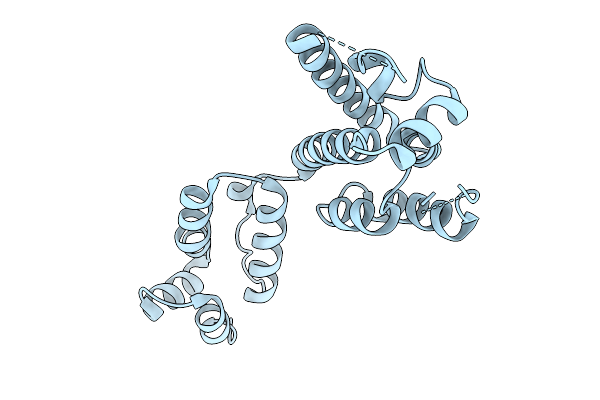
Organism: Faecalibacterium duncaniae
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: TRS, EDO |
 |
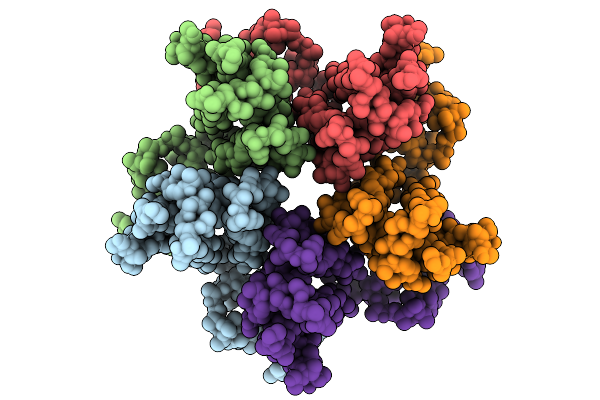
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRUS LIKE PARTICLE |
 |
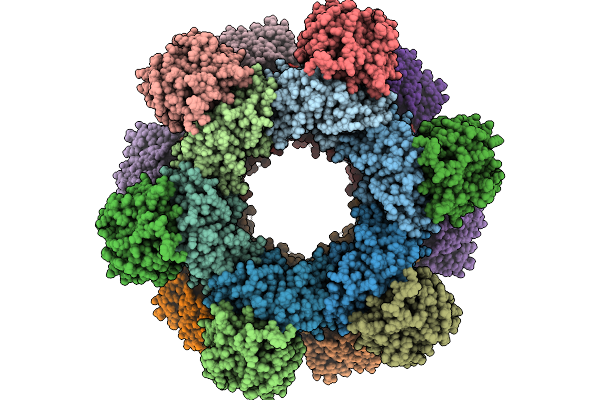
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRUS LIKE PARTICLE |
 |
Cryo-Em Structure Of A Designed Pyridoxal Phosphate (Plp) Synthase Fused To A Designed Circumsporozoite Protein Antigen From Plasmodium Falciparum (Csp-P1-Csp And Csp-P2-Csp)
Organism: Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: BIOSYNTHETIC PROTEIN |
 |
Cryo-Em Structure Of Rnf20/Rnf40-Rad6A-Ub In Complex With H2Bs112Glcnac Nucleosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: NUCLEAR PROTEIN Ligands: ZN, NAG |
 |
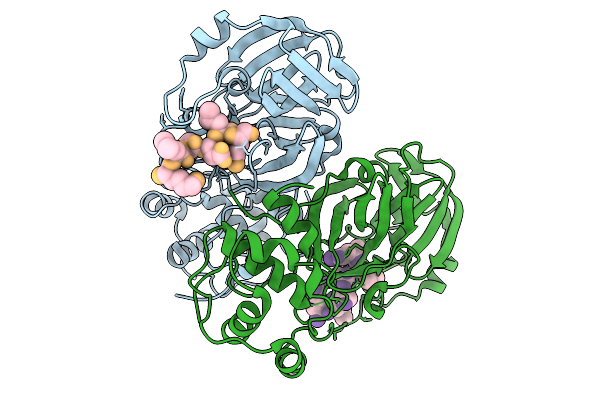
Organism: Porcine epidemic diarrhea virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: Porcine epidemic diarrhea virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN |
 |
Organism: African swine fever virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN |
 |
Organism: Asfarviridae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/DNA Ligands: DCP, MG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: SO4, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: SAH, ZN |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F001
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1JE8, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F005
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PG4, DMS, A1JE7 |
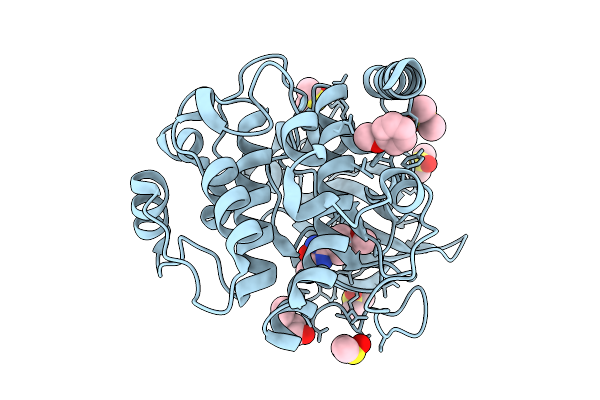 |
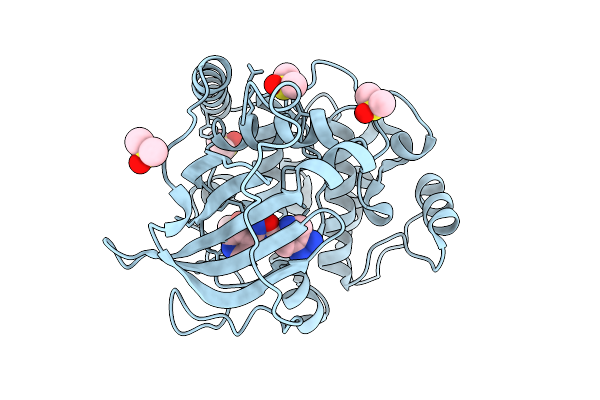
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F009
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE6, PEG |
 |
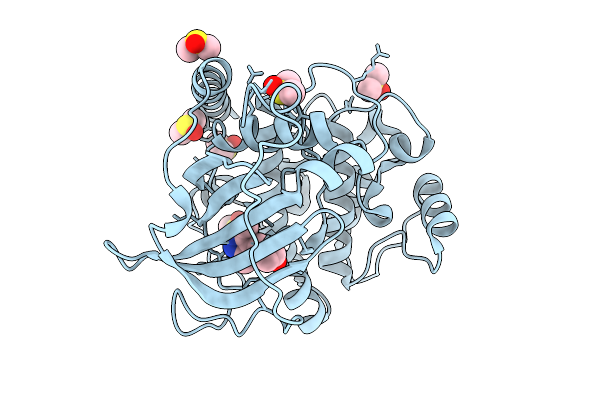
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F012
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE5 |
 |
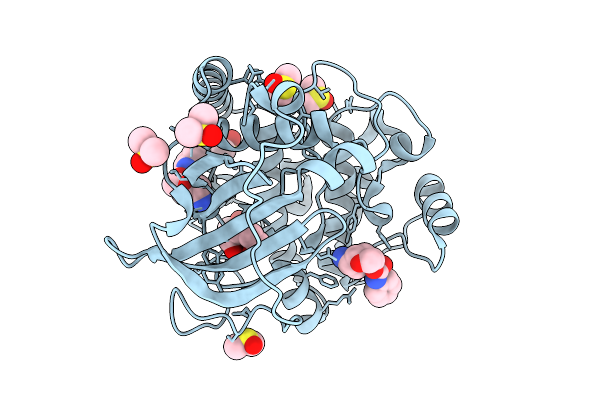
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F024
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE4 |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F030
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PEG, A1JE9, DMS |

