Search Count: 30,548
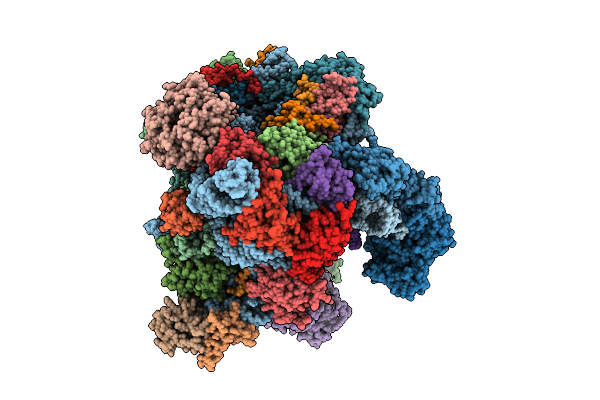 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
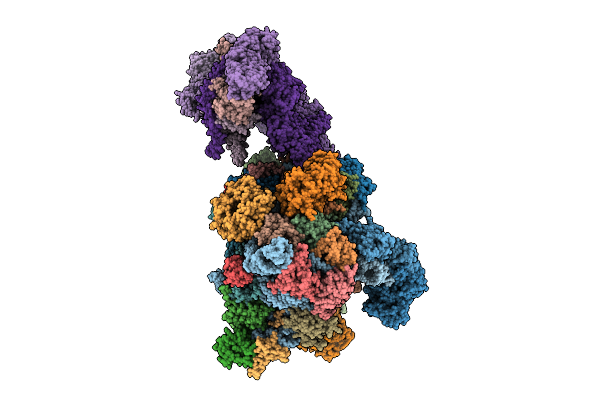 |
Late-Stage 48S Initiation Complex With Eif3 (Ls48S-Eif3 Ic) Guided By The Trans-Rna
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSLATION Ligands: SF4 |
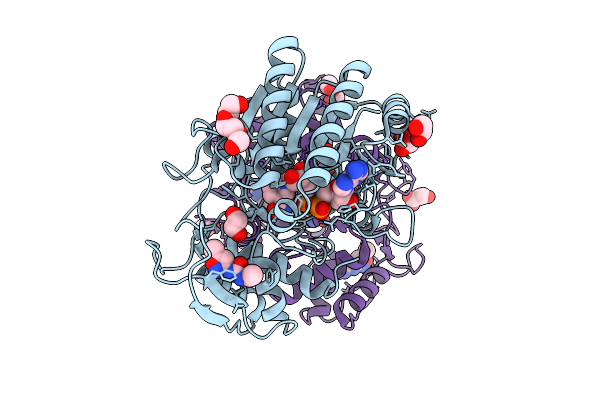 |
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
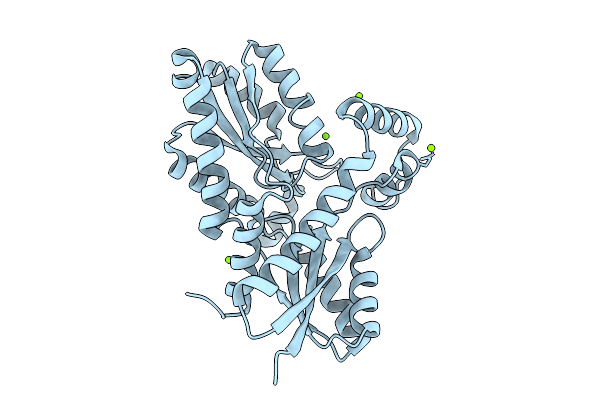 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, MN, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: MG, CL, 9UM, FE2 |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: CO, MG, CL, 9UM |
 |
Ni(Ii)-Bound Bacillus Subtilis Cpfc (Hemh) Y13C Variant Modified With Bromobimane
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, MG, NI, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: 9UM, CU, MG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: ZN, MG, 9UM |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL, PEG |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Streptomyces virginiae
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1L6P, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ISOMERASE Ligands: A1EHL, PE8, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ISOMERASE Ligands: A1EFQ, PE8, SO4 |
 |
Crystal Structure Of Sars-Cov-2 Mpro S10A In Complex With Pfizer Intravenous Inhibitor Pf-00835231
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: V2M |
 |
Crystal Structure Of Sars-Cov-2 Mpro S10C In Complex With Pfizer Intravenous Inhibitor Pf-00835231
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: DMS, EDO, V2M |
 |
Crystal Structure Of Sars-Cov-2 Mpro S113A In Complex With Pfizer Intravenous Inhibitor Pf-00835231
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: V2M |
 |
Crystal Structure Of Sars-Cov-2 Mpro S113C In Complex With Pfizer Intravenous Inhibitor Pf-00835231
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: V2M |

