Search Count: 50
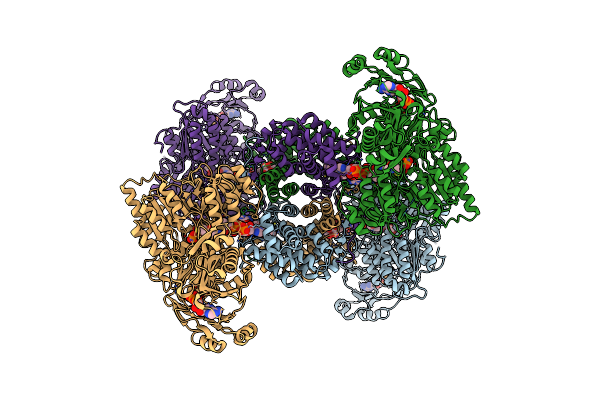 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.40 Å Release Date: 2023-05-10 Classification: LYASE Ligands: ADP, UNL, Q5B, ACO, OAA, PO4 |
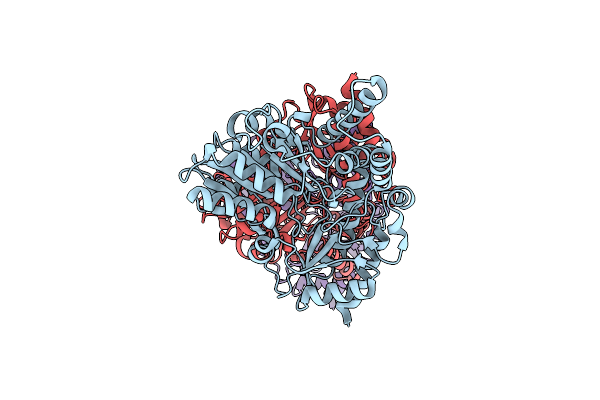 |
Engineered Plp-Dependent Decarboxylative Aldolase From Aspergillus Flavus, Ustd2.0, Bound As The Internal Aldimine
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-05-04 Classification: LYASE, TRANSFERASE |
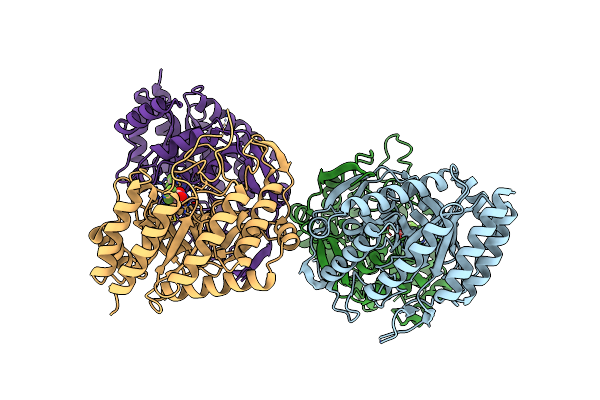 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-11-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 7QE |
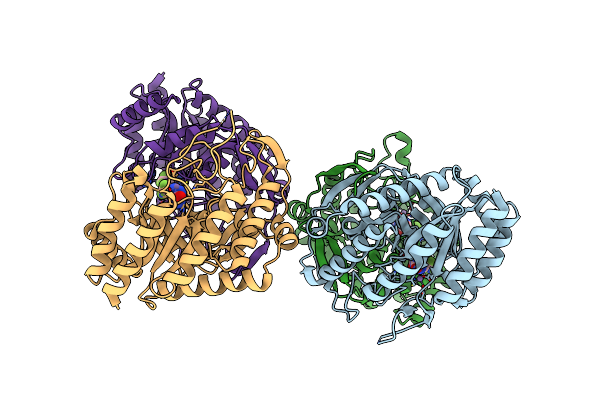 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-11-17 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 7QH, MN |
 |
Crystal Structure Of Catechol O-Methyl Transferase (Comt) From Niastella Koreensis
Organism: Niastella koreensis (strain dsm 17620 / kacc 11465 / nbrc 106392 / gr20-10)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: GOL |
 |
Crystal Structure Of Catechol O-Methyl Transferase (Comt) From Niastella Koreensis
Organism: Niastella koreensis gr20-10
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: SAM |
 |
Crystal Structure Of Catechol O-Methyl Transferase (Comt) From Niastella Koreensis
Organism: Niastella koreensis (strain dsm 17620 / kacc 11465 / nbrc 106392 / gr20-10)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: MG, GOL, SAH |
 |
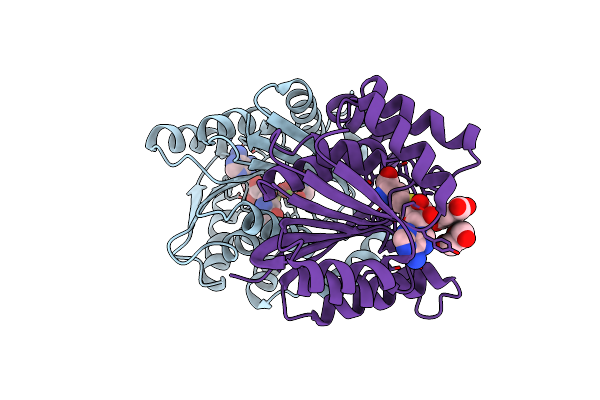
Crystal Structure Of Octaprenyl Diphosphate Synthase From Stenotrophomonas Maltophilia
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Release Date: 2021-02-03 Classification: TRANSFERASE Ligands: CL, EDO |
 |
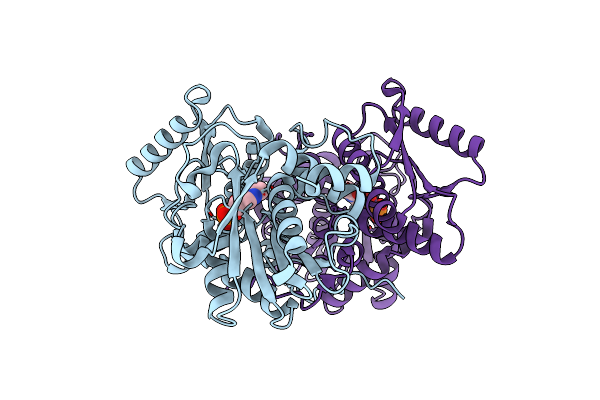
Crystal Structure Of Catechol O-Methyl Transferase (Comt) From Niastella Koreensis
Organism: Niastella koreensis gr20-10
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-18 Classification: TRANSFERASE Ligands: SAH, DHB, MG, GOL |
 |
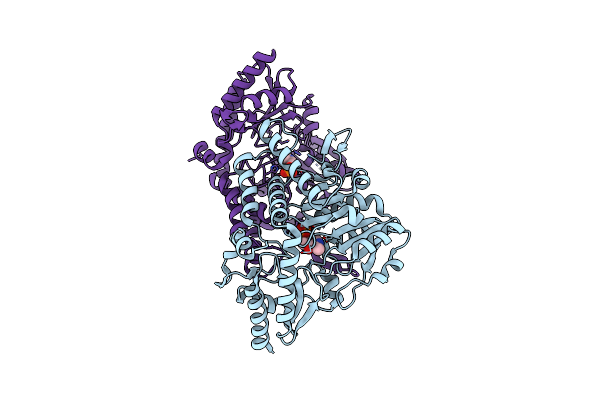
Crystal Structure Of O-Acetyl Serine Sulfhydrylase Isoform 3 From Entamoeba Histolytica
Organism: Entamoeba histolytica
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: PLP |
 |
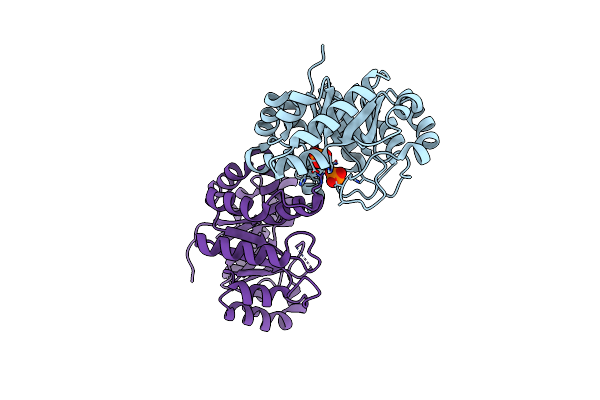
E. Coli Cysteine Desulfurase Sufs With A Spontaneously Rotated Beta-Hairpin
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-03-25 Classification: Lyase, Transferase Ligands: PLP |
 |
Crystal Structure Of A Pyridoxine 5'-Phosphate Synthease From Stenotrophomonas Maltophilia K279A
Organism: Stenotrophomonas maltophilia (strain k279a)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-03-25 Classification: LYASE Ligands: PO4 |
 |
Organism: Escherichia coli (strain k12), Staphylococcus aureus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2017-06-28 Classification: TRANSFERASE |
 |
Mycobacterium Tuberculosis Cysm In Complex With The Urea-Scaffold Inhibitor 3 [4-(3-([1,1'-Biphenyl]-3-Yl)Ureido)-2-Hydroxybenzoic Acid]
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-08-17 Classification: LYASE Ligands: PLP, 6EQ |
 |
Crystal Structure Of Elongation Domain Of Phomopsis Amygdali Fusicoccadiene Synthase
Organism: Phomopsis amygdali
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2016-01-20 Classification: TRANSFERASE |
 |
Crystal Structure Of Elongation Domain Of Phomopsis Amygdali Fusicoccadiene Synthase Complexed With Cobalt Ions And Pamidronate
Organism: Phomopsis amygdali
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-01-20 Classification: TRANSFERASE Ligands: CO, 210 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2015-11-04 Classification: LYASE,TRANSFERASE Ligands: GDP, GOL, MN, ZN, PO4 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-11-04 Classification: LYASE,TRANSFERASE Ligands: GOL, PO4, PEP |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2015-11-04 Classification: LYASE,TRANSFERASE Ligands: GDP, GOL, PEP |
 |
Crystal Structure Of E83A Mutant Of Mtb Pepck In Complex With Zn2+ And Phosphate Ion
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-11-04 Classification: LYASE,TRANSFERASE Ligands: GOL, ZN, PO4 |

