Search Count: 9
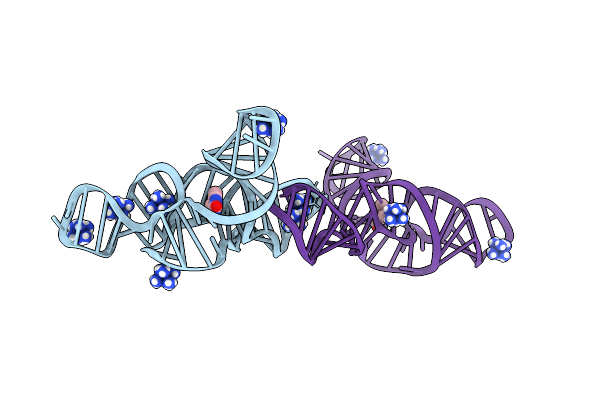 |
Structure Of The Bacillus Subtilis Yjdf Riboswitch Complexed With Lumichrome In The Presence Of Iridium Hexammine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-11-19 Classification: RNA Ligands: LUM, IRI |
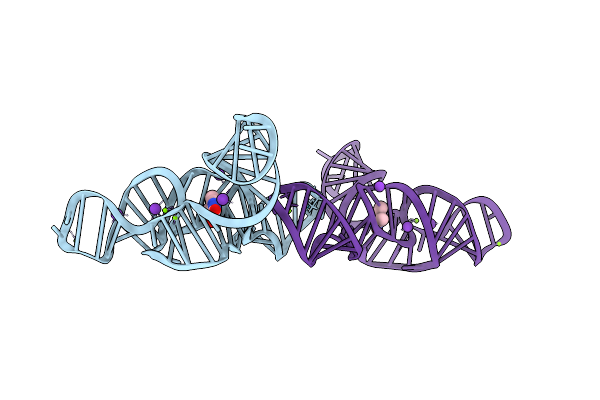 |
Structure Of The Bacillus Subtilis Yjdf Riboswitch Aptamer Domain In Complex With Lumichrome
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-11-19 Classification: RNA Ligands: MG, K, LUM |
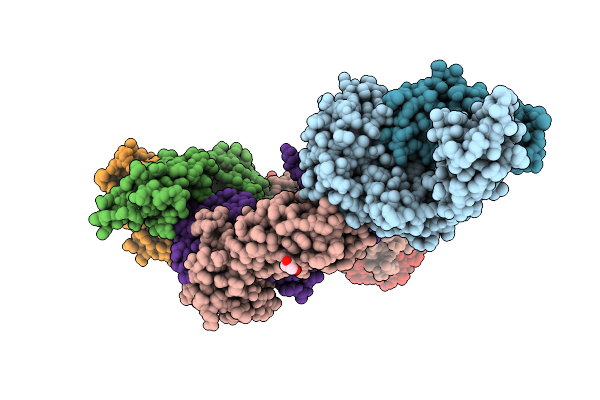 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: GOL, LUM, CL |
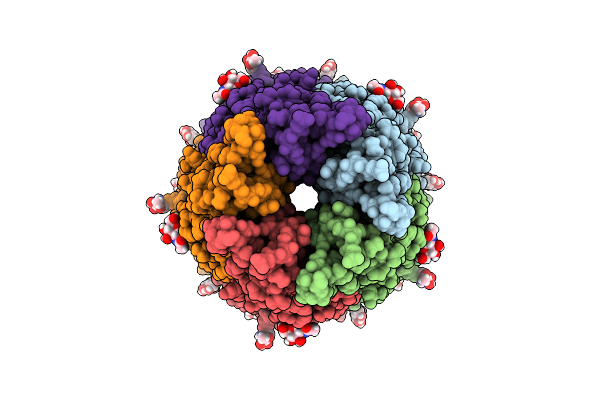 |
Organism: Octopus bimaculoides
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: NAG, LUM |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-02-27 Classification: FLAVOPROTEIN Ligands: FMN, MG, CL, LUM |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2006-02-15 Classification: FLAVOPROTEIN Ligands: MG, NA, CL, SO4, LUM |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-02-01 Classification: FLAVOPROTEIN Ligands: MG, NA, CL, SO4, LUM |
 |
Crystal Structure Of Flavin Binding To Fad Synthetase From Thermotoga Maritina
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2004-10-19 Classification: TRANSFERASE Ligands: MG, LUM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2001-02-28 Classification: BILIVERDIN-IX BETA REDUCTASE Ligands: NAP, LUM |

