Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Corynebacterium glutamicum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSPORT PROTEIN Ligands: PGT, ABU, LMT |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The K2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The O2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
Organism: Erythrobacter sanguineus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: BCL, A1EL2, A1ELD, 8K6, PGV, LMT, BPH, UQ8, FE, H4X, PEF, PO4, LHG |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSPORT PROTEIN Ligands: LMT |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: Y01, MN, LMT |
 |
Cryo Em Structure Of Rc-Dlh Complex Model Ii From Gemmatimonas Groenlandica
Organism: Gemmatimonas groenlandica
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, LMT, MQ8, FE, CD4, CRT, PEX, HEC, V7N |
 |
Structure Of The Double Cys-Substituted Cross-Linked Acrb Variant S562C_T837C
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: LMT, D10, GOL, D12, HEX, C14, OCT, DD9, SO4 |
 |
Cryo-Em Structure Of The Monomeric Rhodobacter Sphaeroides G1C Lh1-Rc Core Complex
Organism: Cereibacter sphaeroides
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: PHOTOSYNTHESIS Ligands: BCL, BPH, U10, PGV, LDA, LMT, FE, CDL, A1EF2 |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: LMT, OCT, C14, EDO, GOL, D12, HEX, XE9, D10, DDR, DDQ, SO4, LPX |
 |
Cryo-Em Structure Of The Wild-Type Human Serotonin Transporter Complexed With S-Ketamine
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: CL, NA, Y01, LMT, NAG, JC9, DD9, D12, D10, OCT |
 |
Cryo-Em Structure Of The Apo-Form Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
 |
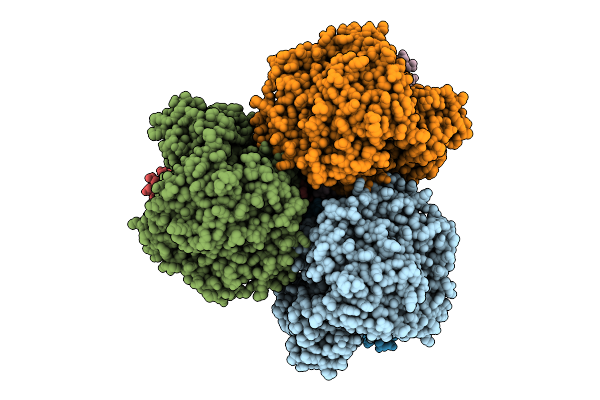
Cryo-Em Structure Of The Lipid-Bound Succiante Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SIN, SF4, FES, F3S, HEM, LMT, PGV, PEV |
 |
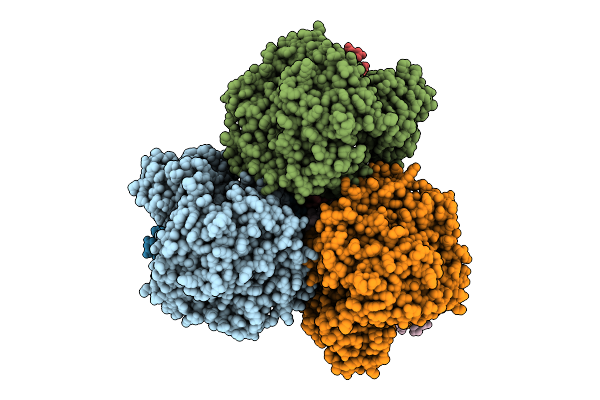
Cryo-Em Structure Of The Mk7-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, MQ7 |
 |
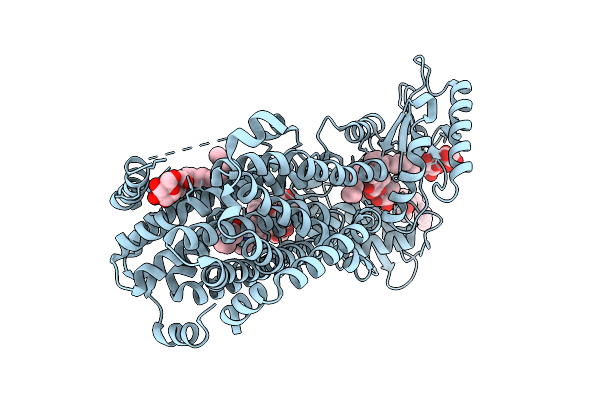
Cryo-Em Structure Of The Mk4-Bound Succinate Dehydrogenase From Chloroflexus Aurantiacus
Organism: Chloroflexus aurantiacus j-10-fl
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: FAD, SF4, FES, F3S, HEM, LMT, 1L3 |
 |
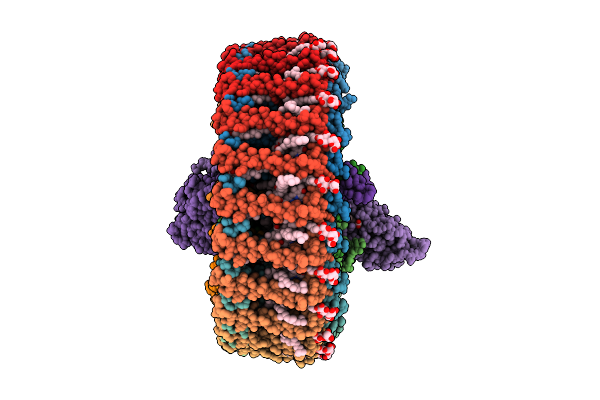
Crystal Structure Of Mycolic Acid Transporter Mmpl3 From Mycobacterium Smegmatis Complexed With Indolcarboxamide
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: A1L5O, LMT |
 |
Cryo-Em Structure Of Rc-Dlh Complex Model I From Gem. Groenlandica Strain Tet16
Organism: Gemmatimonas groenlandica
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PHOTOSYNTHESIS Ligands: V7N, BCL, LMT, PEX, MQ8, CD4, BPH, FE, CRT, HEC |
 |
Organism: Chroococcidiopsis thermalis pcc 7203
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: CL0, F6C, CLA, PQN, SF4, BCR, LHG, LMT, LMG, LFA, CA |
 |
Cryo-Em Structure Of A Tri-Heme Cytochrome-Associated Rc-Lh1 Complex From A Marine Photoheterotrophic Bacterium, Purified With Edta-Containing Solutions
Organism: Dinoroseobacter shibae dfl 12 = dsm 16493
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: BCL, SPN, MW9, LMT, FE, BPH, U10, CDL, HEC |