Search Count: 110
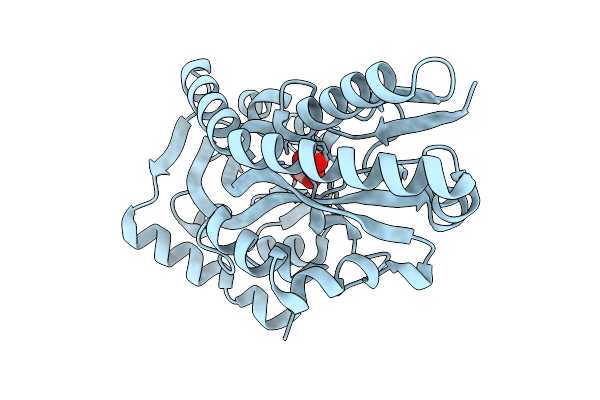 |
Crystal Structure Of C4-Dicarboxylate-Binding Protein (Pa0884) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With L-Malate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: LMR |
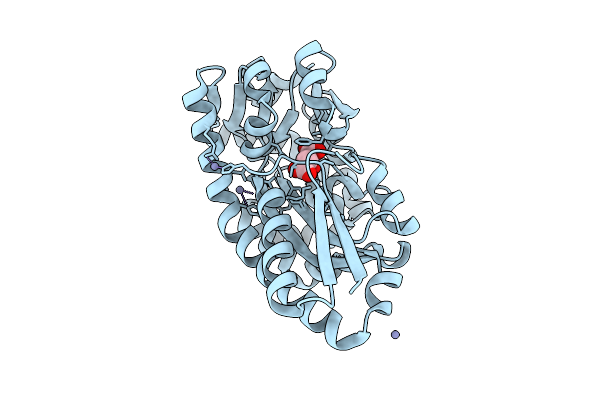 |
Crystal Structure Of C4-Dicarboxylate-Binding Periplasmic Protein (Pa5167) Of Tripartite Atp-Independent Periplasmic Transporter Family From Pseudomonas Aeruginosa Pao1 In Complex With L-Malate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-07-09 Classification: TRANSPORT PROTEIN Ligands: LMR, ZN |
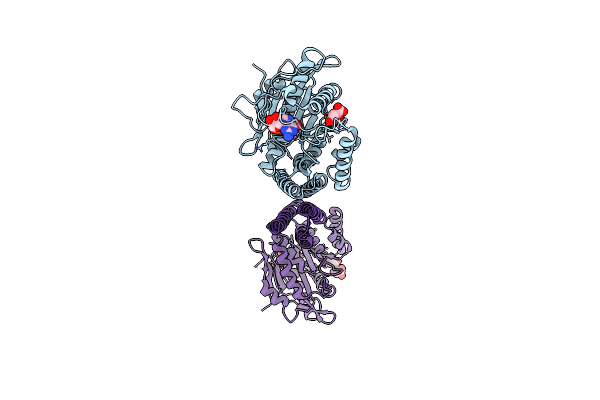 |
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2025-04-23 Classification: HYDROLASE Ligands: GDP, LMR |
 |
Arabidopsis Thaliana Phosphoenolpyruvate Carboxylase Ppc1 R886G Mutant With Bound Malate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2024-10-30 Classification: PLANT PROTEIN Ligands: LMR, CL, PEG |
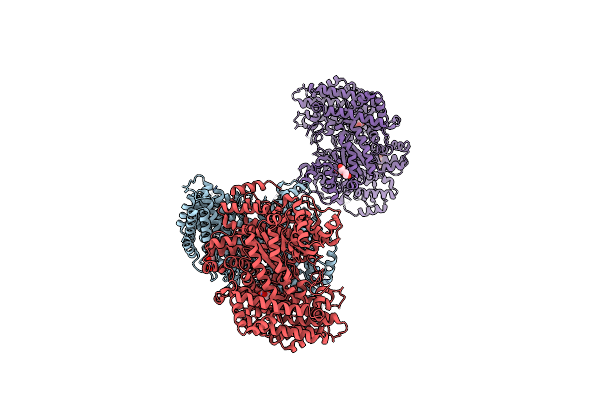 |
Arabidopsis Thaliana Phosphoenolpyruvate Carboxylase Ppc1 G678S Mutant With Bound Malate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2024-10-23 Classification: PLANT PROTEIN Ligands: LMR, CL, PEG |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: LMR, GOL, NAD |
 |
Organism: Anabaena sp. 90
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: LMR, NAD |
 |
Preclinical Characterization Of Pan-Nkg2D Ligand-Binding Nkg2D Receptor Decoys
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-04-17 Classification: IMMUNE SYSTEM Ligands: LMR, NAG, GOL |
 |
Arabidopsis Thaliana Phosphoenolpyruvate Carboxylase Ppc1 T778 Mutant With Bound Malate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-04-03 Classification: PLANT PROTEIN Ligands: LMR, CL, PEG |
 |
Arabidopsis Thaliana Phosphoenolpyruvate Carboxylase Ppc1 Mutant A651V In Complex With L-Malate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2024-02-21 Classification: PLANT PROTEIN Ligands: LMR, CL |
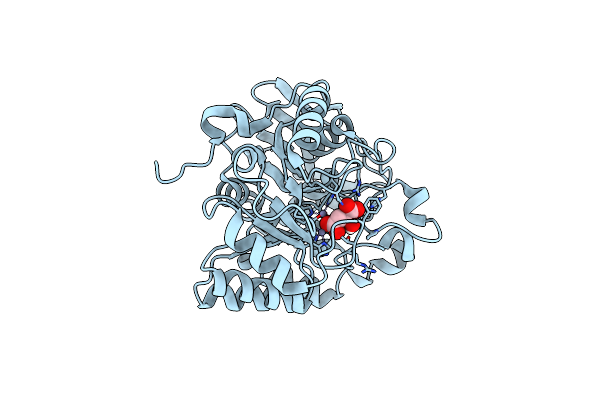 |
Crystal Structure Of The Human Dihydroorotase Domain In Complex With Malic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-09-20 Classification: HYDROLASE Ligands: LMR, ZN |
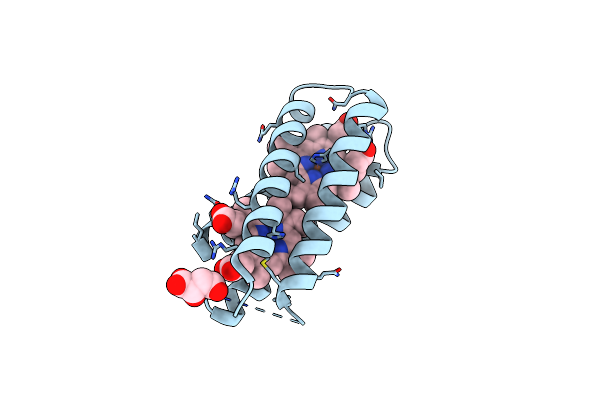 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-08-09 Classification: ELECTRON TRANSPORT Ligands: HEM, LMR, CL |
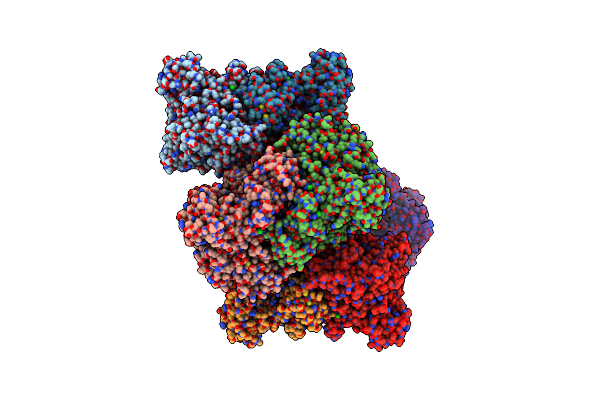 |
Arabidopsis Thaliana Phosphoenolpyruvate Carboxylase Ppc1 In Complex With L-Malate
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2023-05-31 Classification: PLANT PROTEIN Ligands: LMR, PEG, CL |
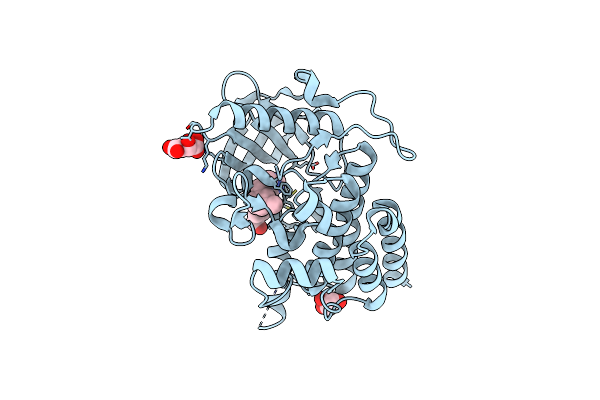 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-05-24 Classification: LIGASE Ligands: LMR, GOL, UIK |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-05-17 Classification: TRANSPORT PROTEIN Ligands: 8XI, MLI, LMR, FMT, ACT |
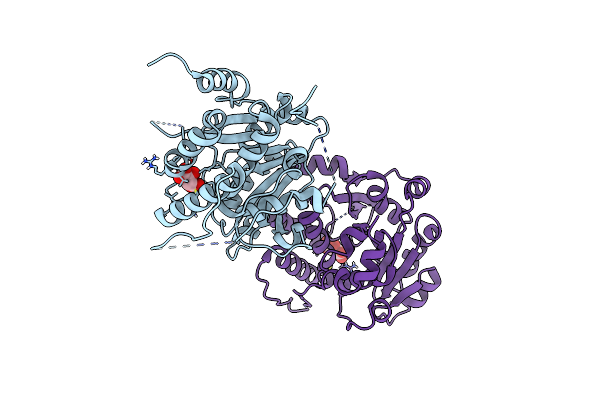 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.1 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: LMR, MLT |
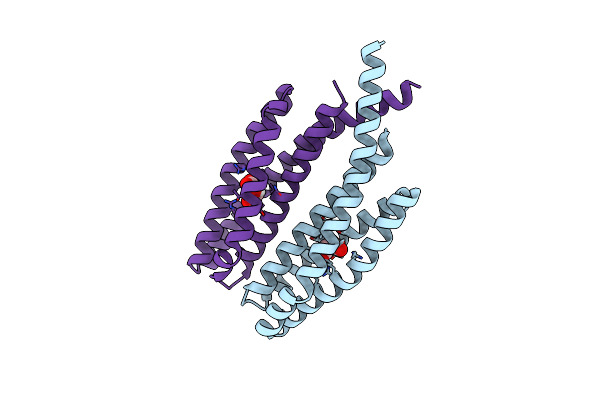 |
Organism: Comamonas testosteroni cnb-2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-01-04 Classification: SIGNALING PROTEIN Ligands: LMR |
 |
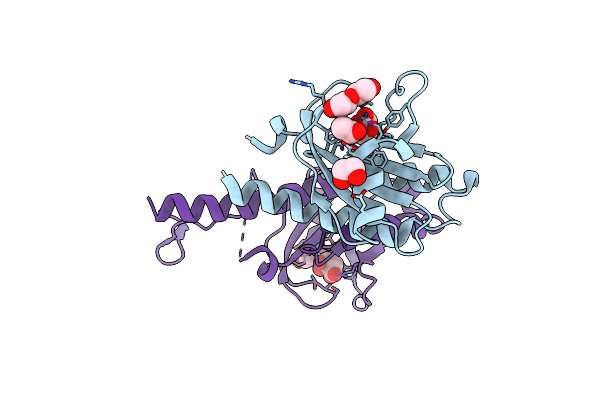
X-Ray Structure Of The Complex Between Human Alpha Thrombin And A Pseudo-Cyclic Thrombin Binding Aptamer (Tba-Nnp/Ddp) - Crystal Form Alpha
Organism: Synthetic construct, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2022-11-30 Classification: HYDROLASE Ligands: 0G6, NA, LMR, JL0, JKR, K |
 |
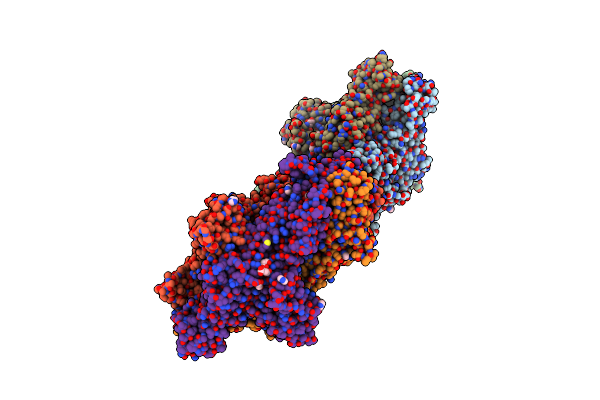
The Novel Membrane-Proximal Sensing Mechanism In A Broad-Ligand Binding Chemoreceptor Mcpa Of Bacillus Velezensis
Organism: Bacillus velezensis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-07-06 Classification: PROTEIN BINDING Ligands: LMR, EDO, ACT, PEG |
 |
Crystal Structure Of Klebsiella Pneumoniae K1 Capsule-Specific Polysaccharide Lyase In A P1 Crystal Form
Organism: Klebsiella phage ntuh-k2044-k1-1
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-05-18 Classification: VIRAL PROTEIN Ligands: LMR, IMD, GOL |

