Search Count: 165
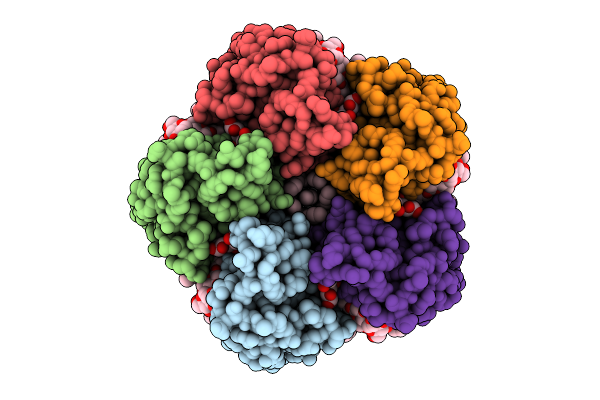 |
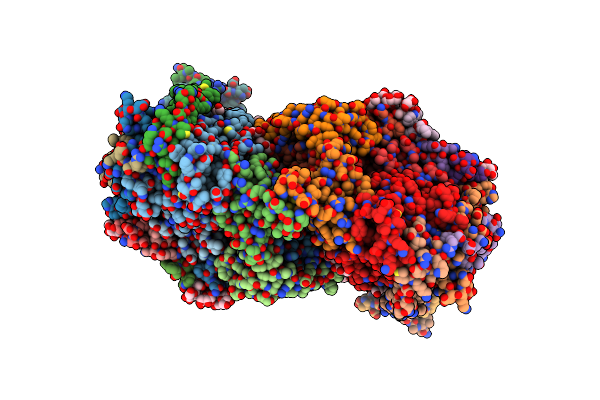
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
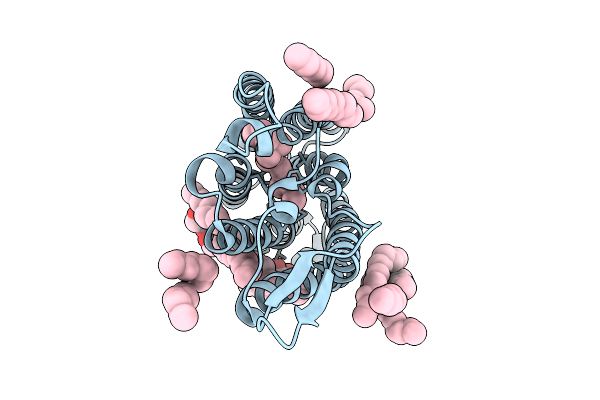 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The K2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
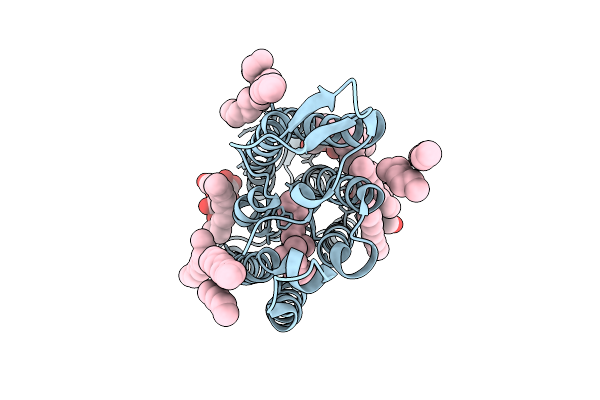 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The O2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
 |
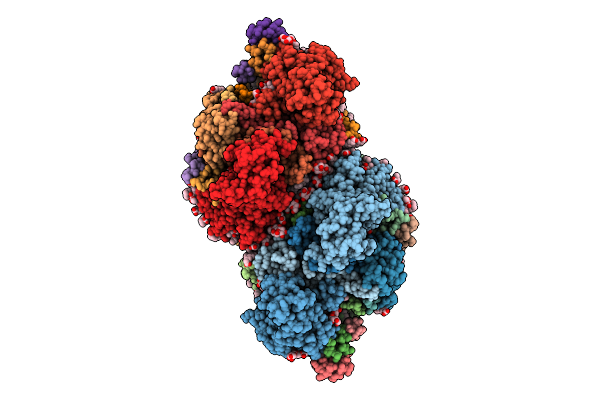
Organism: Chroococcidiopsis thermalis pcc 7203
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: PHOTOSYNTHESIS Ligands: CL0, F6C, CLA, PQN, SF4, BCR, LHG, LMT, LMG, LFA, CA |
 |
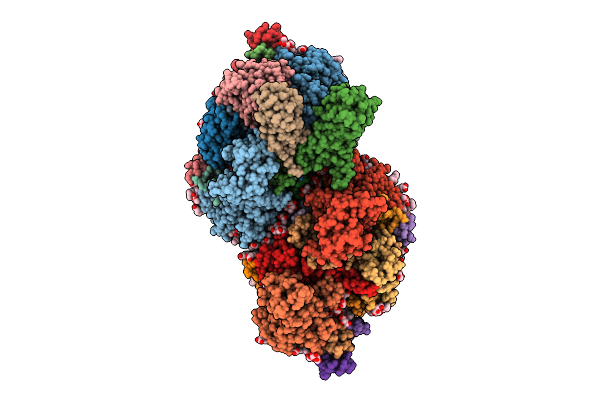
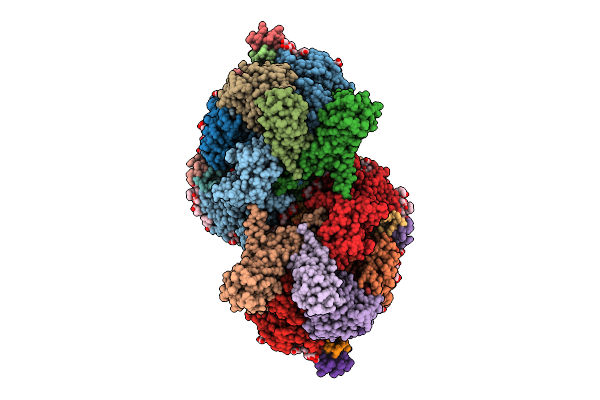
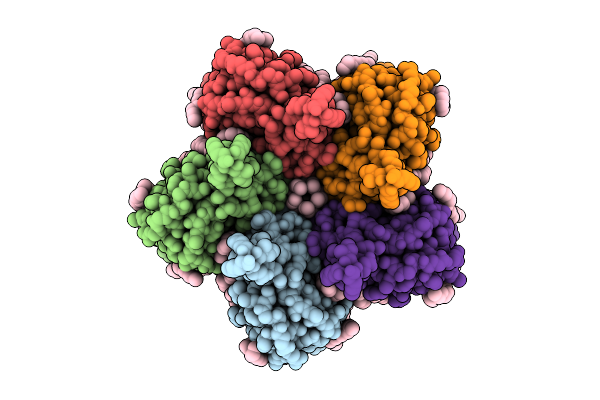
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PHOTOSYNTHESIS Ligands: FE2, CL, CLA, PHO, BCR, SQD, PL9, LFA, PLM, BCT, LHG, LMG, LMT, DGD, HEM, RRX |
 |
 |
 |
Cryo-Em Structure Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA |
 |
Cryo-Em Structure Of The Light-Driven Proton Pump Pspr In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET, LMT |
 |
Cryo-Em Structure Of The Double Mutant H84V/E120G Of The Flotillin-Associated Rhodopsin Psfar In Detergent Micelle
Organism: Candidatus pseudothioglobus sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Room Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 95% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Room-Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 85% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Bovine Heart Cytochrome C Oxidase In The Carbon Dioxide-Bound Fully Oxidized State
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, CDL, CO2, LFA, DMU, EDO, PGV, CUA, CHD, UNX, ZN, PEK |
 |
Bovine Heart Cytochrome C Oxidase In The Carbon Dioxide-Bound Fully Reduced State
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, CO2, LFA, DMU, EDO, PGV, CUA, CHD, UNX, CDL, ZN, PEK |
 |
Bovine Heart Cytochrome C Oxidase In The Nitrous Oxide-Bound Fully Oxidized State
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, N2O, LFA, DMU, EDO, PGV, CUA, CHD, UNX, CDL, ZN, PEK |
 |
Bovine Heart Cytochrome C Oxidase In The Nitrous Oxide-Bound Fully Reduced State
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, N2O, LFA, DMU, EDO, PGV, CUA, CHD, UNX, CDL, ZN, PEK |
 |
Bovine Heart Cytochrome C Oxidase In The Xenon-Bound Fully Oxidized State Under Aerobic Condition
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, CDL, LFA, DMU, EDO, XE, PGV, CUA, CHD, UNX, ZN, PEK |
 |
Bovine Heart Cytochrome C Oxidase In The Xenon-Bound Fully Oxidized State Under Anaerobic Condition
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, CDL, LFA, DMU, EDO, XE, PGV, CUA, CHD, UNX, ZN, PEK |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, CDL, LFA, DMU, EDO, XE, PGV, CUA, CHD, UNX, ZN, PEK |