Search Count: 93
 |
Human Glyoxalase I (With C-Ter His Tag) In Complex With Licochalcone B, Form 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LYASE Ligands: ZN, A1L5R |
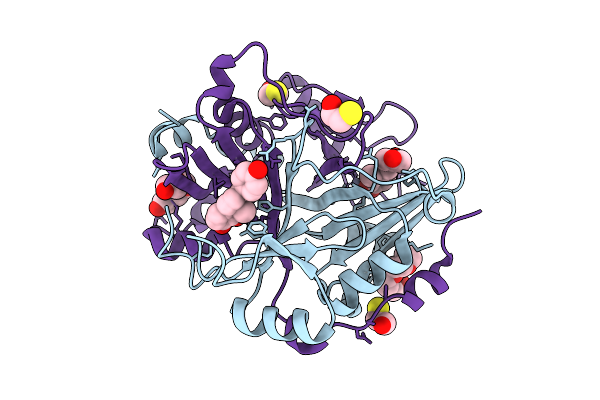 |
Human Glyoxalase I (With C-Ter His Tag) In Complex With Licochalcone B, Form 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LYASE Ligands: A1L5R, PG4, BME, ZN |
 |
Human Glyoxalase I (With C-Ter His Tag) In Complex With Licochalcone B And Gsh, Form 3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LYASE Ligands: ZN, A1L5R, PG4, GSH |
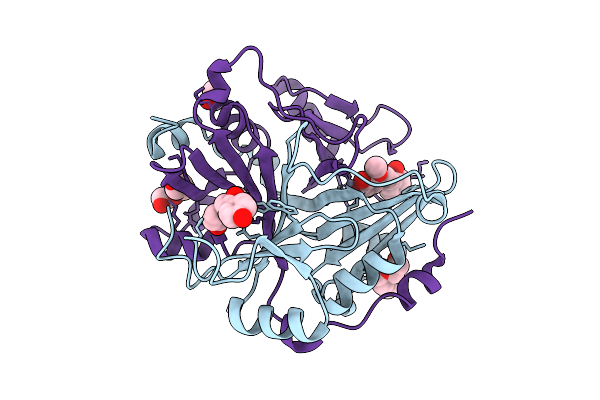 |
Human Glyoxalase I (With C-Ter His Tag) In Complex With Licochalcone B, Form 4
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LYASE Ligands: A1L5R, PGE, ZN, BME |
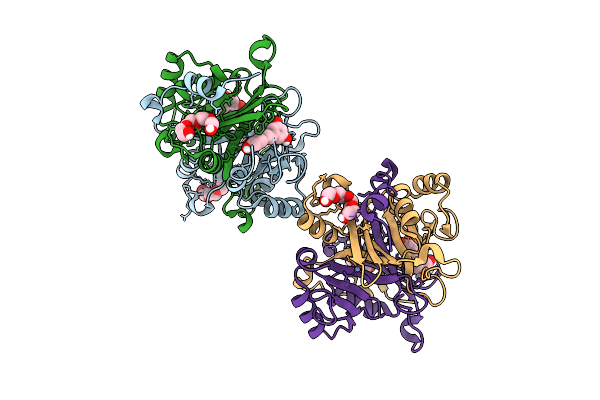 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LYASE Ligands: ZN, PIT, PG4 |
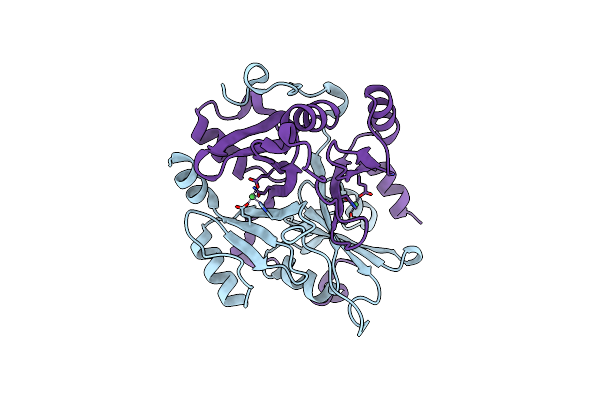 |
Organism: Gossypium hirsutum
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-07-27 Classification: LYASE Ligands: NI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-04-13 Classification: LYASE Ligands: ZN, GOL, BO3, IMD, BME |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-13 Classification: LYASE Ligands: ZN, 5ZO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2022-04-13 Classification: LYASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-13 Classification: LYASE Ligands: ZN, 5ZO |
 |
Crystal Structure Of Mouse Glyoxalase I Complexed With A Small Molecule Inhibitor
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-09-30 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, E1L |
 |
Crystal Structure Of V278E-Glyoxalase I Mutant From Zea Mays In Space Group P4(1)2(1)2
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: GSH, CO, FMT |
 |
Crystal Structure Of V278E-Glyoxalase I Mutant From Zea Mays In Space Group P6(3)
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: CO |
 |
Crystal Structure Of E144Q-Glyoxalase I Mutant From Zea Mays In Space Group P4(1)2(1)2
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-11-21 Classification: PLANT PROTEIN Ligands: CO, GSH, FMT |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-09-16 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, 3WL |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-09-09 Classification: LYASE Ligands: NI, FMT, NA, 2PE |
 |
Crystal Structure Of A Probable Lactoylglutathione Lyase From Brucella Melitensis In Complex With Glutathione
Organism: Brucella abortus (strain 2308)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2015-04-22 Classification: LYASE Ligands: GSH, ZN, MRD, MPD |
 |
Crystal Structure Of Mouse Glyoxalase I In Complexed With 18-Beta-Glycyrrhetinic Acid
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-03-18 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, CBW |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-02-11 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, ZBF |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-10-15 Classification: LYASE |

