Search Count: 35
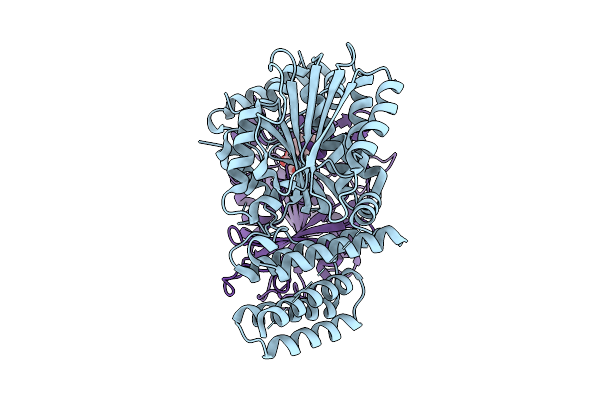 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: HORMONE Ligands: GA3 |
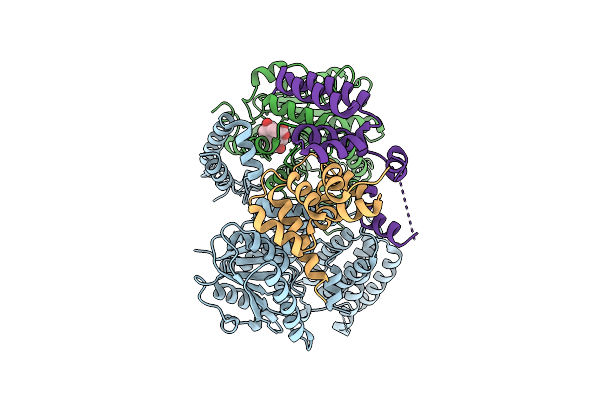 |
Cryo-Em Structure Of Arabidopsis Thaliana Rga In Complex With Gid1A, Sly1, And Ask2 (Consensus Map)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: HORMONE Ligands: GA3 |
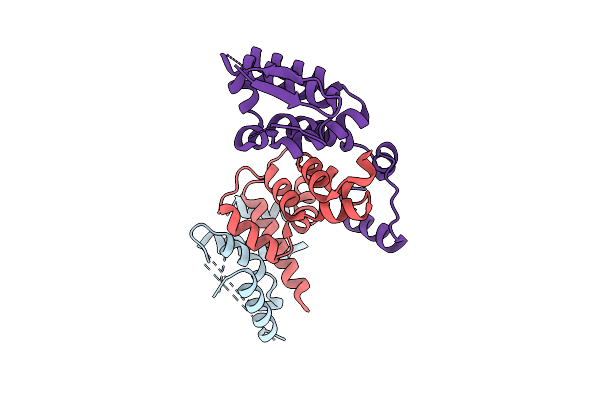 |
Cryo-Em Structure Of Arabidopsis Thaliana Rga In Complex With Gid1A, Sly1, And Ask2 (Focused Map)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: HORMONE |
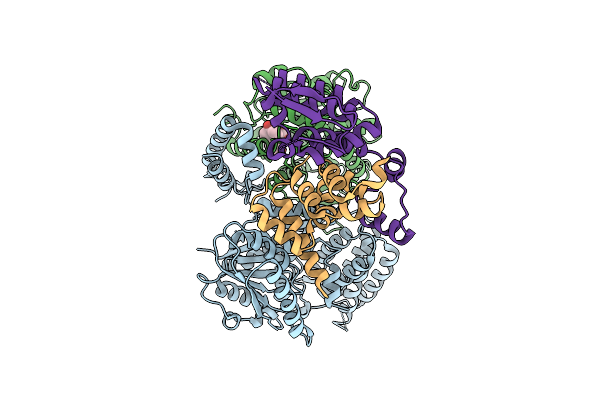 |
Cryo-Em Structure Of Arabidopsis Thaliana Rga In Complex With Gid1A, Sly1, And Ask2 (Composite Map)
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: HORMONE Ligands: GA3 |
 |
Crystal Structure Of Bacillus Subtilis Glyceraldehyde-3-Phosphate Dehydrogenase Gapb
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-07-03 Classification: HYDROLASE Ligands: CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-07-03 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-05-08 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-05-08 Classification: TRANSFERASE Ligands: ZN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-04-19 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-04-05 Classification: ANTIMICROBIAL PROTEIN/INHIBITOR |
 |
Crystal Structure Of Bacillus Subtilis Lyte Catalytic Domain In Complex With Isea
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2023-04-05 Classification: ANTIMICROBIAL PROTEIN/INHIBITOR |
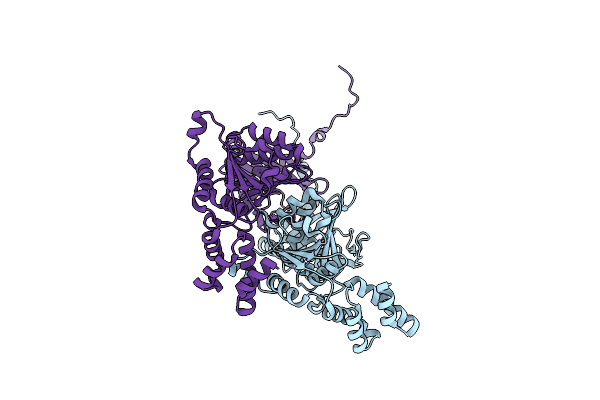 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2022-10-19 Classification: HYDROLASE |
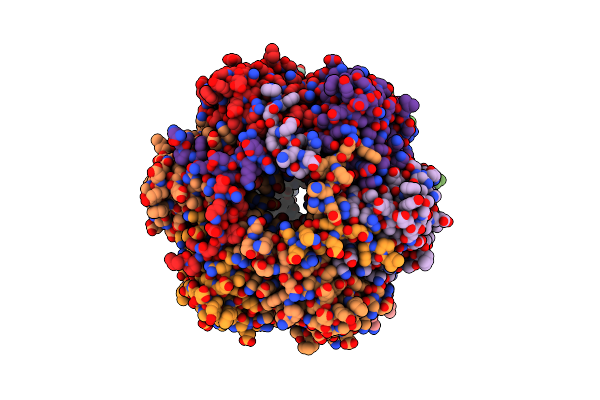 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-10-19 Classification: HYDROLASE |
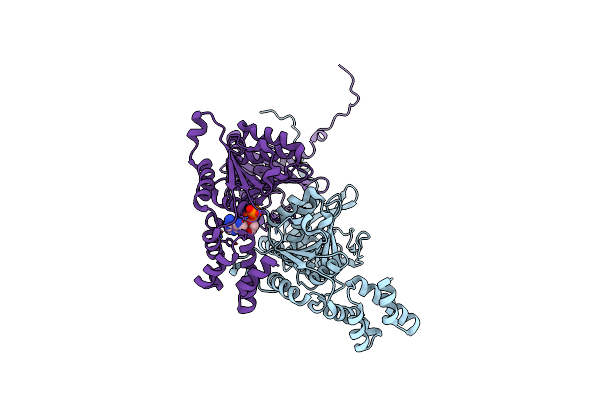 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-10-19 Classification: HYDROLASE Ligands: ADP |
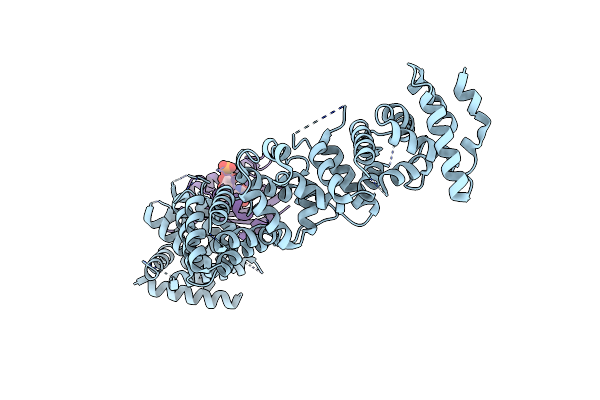 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2022-04-27 Classification: CIRCADIAN CLOCK PROTEIN Ligands: FMN |
 |
Crystal Structure Of The Tandem B-Box Domain Of Arabidopsis Thaliana Constans
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-02 Classification: PLANT PROTEIN Ligands: ZN |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2021-04-21 Classification: DNA BINDING PROTEIN |
 |
The Crystal Structure Of B. Subtilis Rsbv/Rsbw Complex In The Monoclinic Crystal Form
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-07-15 Classification: SIGNALING PROTEIN |
 |
The Crystal Structure Of B. Subtilis Rsbv/Rsbw Complex In The Hexagonal Crystal Form
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-07-15 Classification: SIGNALING PROTEIN |

