Search Count: 47
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2021-05-05 Classification: TRANSFERASE Ligands: NAG, UDP, TRS |
 |
Crystal Structure Of Pomgnt2 In Complex With Udp And Mono-Mannosyl Peptide (379Man Long Peptide)
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2021-05-05 Classification: TRANSFERASE Ligands: NAG, UDP, TRS, MAN |
 |
Crystal Structure Of Pomgnt2 In Complex With Udp And Mono-Mannosyl Peptide (379Man Short Peptide)
Organism: Bos taurus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-05-05 Classification: TRANSFERASE Ligands: NAG, UDP, TRS, MAN |
 |
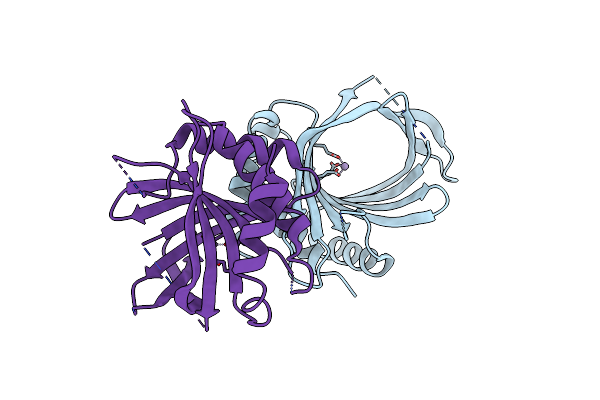
Crystal Structure Of Cet1 From Trypanosoma Cruzi In Complex With Tripolyphosphate, Manganese And Iodide Ions.
Organism: Trypanosoma cruzi strain cl brener
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: MN, 3PO, IOD |
 |
Crystal Structure Of Cet1 From Trypanosoma Cruzi In Complex With Manganese Ion.
Organism: Trypanosoma cruzi strain cl brener
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: MN |
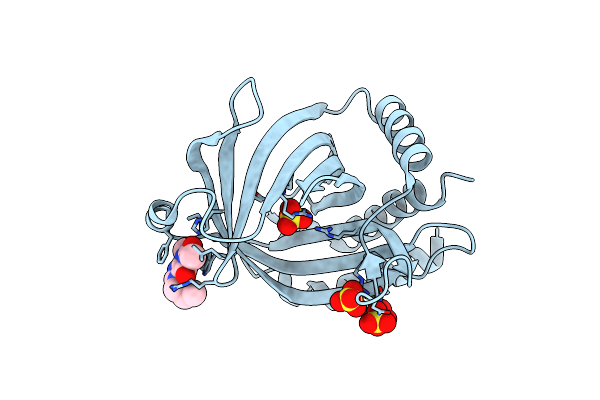 |
Crystal Structure Of Cet1 From Trypanosoma Cruzi In Complex With #951 Ligand
Organism: Trypanosoma cruzi strain cl brener
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: E7R, SO4 |
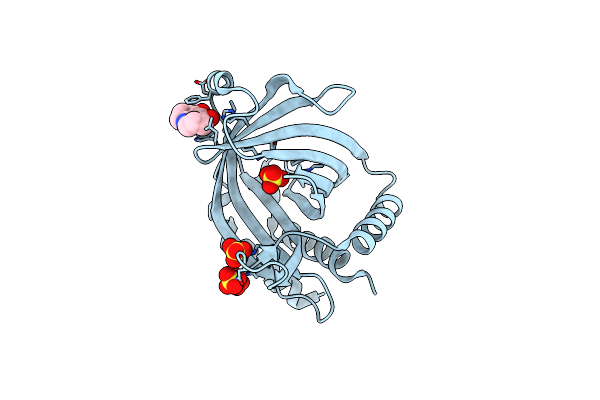 |
Crystal Structure Of Cet1 From Trypanosoma Cruzi In Complex With #466 Ligand.
Organism: Trypanosoma cruzi strain cl brener
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2020-06-03 Classification: HYDROLASE Ligands: JJY, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2020-01-29 Classification: TRANSFERASE Ligands: ZN, NAG, CDP, RB0, BA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, C5P, MG |
 |
Crystal Structure Of Fkrp In Complex With Ba Ion, Cdp-Ribtol, And Sugar Acceptor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, CDP, RB0, BA, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, BA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, MG |
 |
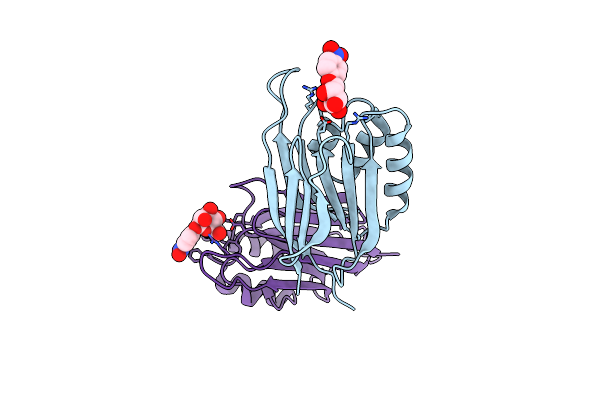
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2020-01-15 Classification: TRANSFERASE Ligands: ZN, NAG, BA |
 |
Serial Femtosecond X-Ray Structure Of A Stem Domain Of Human O-Mannose Beta-1,2-N-Acetylglucosaminyltransferase Solved By Se-Sad Using Xfel (Refined Against 13,000 Patterns)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-08-30 Classification: SUGAR BINDING PROTEIN Ligands: MBE |
 |
Serial Femtosecond X-Ray Structure Of Agrocybe Cylindracea Galectin With Lactose Solved By Se-Sad Using Xfel (Refined Against 60,000 Patterns)
Organism: Agrocybe cylindracea
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: SUGAR BINDING PROTEIN |
 |
Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
Organism: Photinus pyralis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: HYDROLASE Ligands: HG, MG, MPD |
 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2016-12-07 Classification: TRANSCRIPTION Ligands: DLC |
 |
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2016-12-07 Classification: TRANSCRIPTION Ligands: VDP |

