Search Count: 13
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadp And Malonate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: NAP, MLA, NA |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Citrate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadph And Oxalate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: OXL, NDP, GOL, CL, NA |
 |
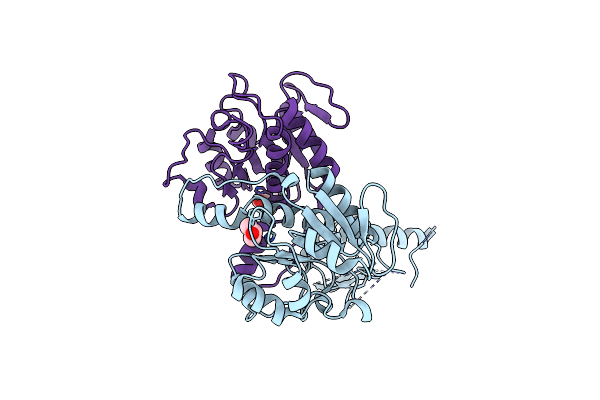
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With Nadp And 2-Keto-D-Gluconic Acid
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-03-29 Classification: OXIDOREDUCTASE Ligands: NAP, 8YV, NA, PEG, CL, PG4, MG, GOL |
 |
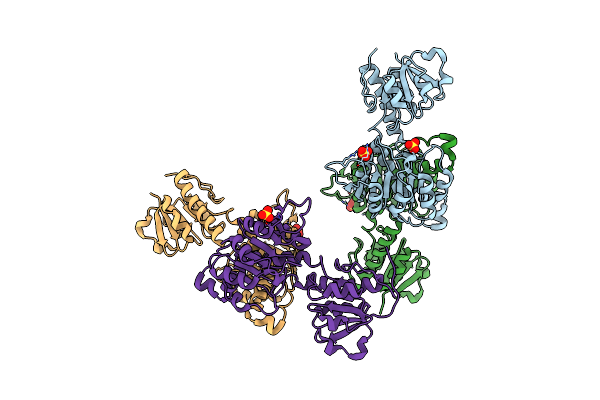
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc02828 (Smghra) From Sinorhizobium Meliloti In Apo Form
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: GOL, CL |
 |
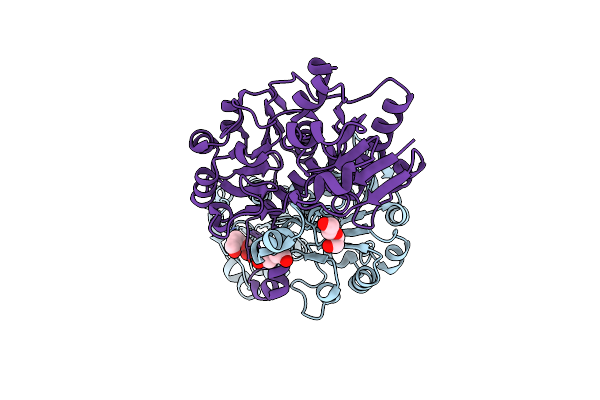
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Apo Form
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-02-22 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
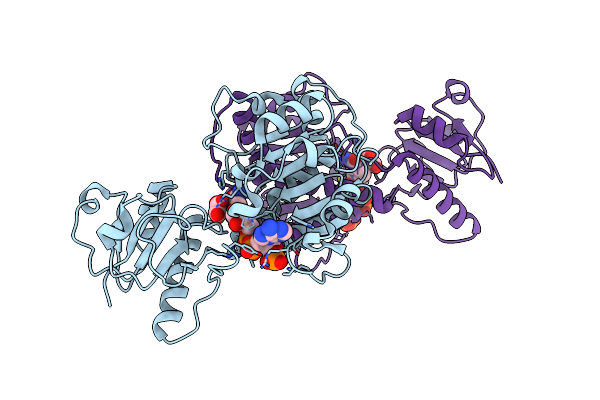
Crystal Structure Of D-Isomer Specific 2-Hydroxyacid Dehydrogenase From Desulfovibrio Vulgaris
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2016-12-07 Classification: OXIDOREDUCTASE Ligands: PEG, PGE |
 |
Crystal Structure Of Nadph-Dependent 2-Hydroxyacid Dehydrogenase From Rhizobium Etli Cfn 42 In Complex With Nadph And Oxalate
Organism: Rhizobium etli (strain cfn 42 / atcc 51251)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-11-16 Classification: OXIDOREDUCTASE Ligands: NDP, OXD |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc04462 (Smghrb) From Sinorhizobium Meliloti In Complex With 2'-Phospho-Adp-Ribose
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-04-13 Classification: OXIDOREDUCTASE Ligands: ACT, CL, A2R |
 |
Crystal Structure Of Sam-Dependent Methyltransferase From Thiobacillus Denitrificans In Complex With S-Adenosyl-L-Homocysteine
Organism: Thiobacillus denitrificans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-11-25 Classification: TRANSFERASE Ligands: SAH, NA |
 |
Crystal Structure Of Sam-Dependent Methyltransferase From Geobacter Sulfurreducens In Complex With S-Adenosyl-L-Homocysteine
Organism: Geobacter sulfurreducens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-06-10 Classification: TRANSFERASE Ligands: SAH, CL |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc02828 (Smghra) From Sinorhizobium Meliloti In Complex With Nadph And Oxalate
Organism: Rhizobium meliloti (strain 1021)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-04-08 Classification: OXIDOREDUCTASE Ligands: CL, OXD, EPE, GOL, NDP |
 |
Crystal Structure Of Nadph-Dependent Glyoxylate/Hydroxypyruvate Reductase Smc02828 (Smghra) From Sinorhizobium Meliloti In Complex With Nadp And Sulfate
Organism: Rhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-09-24 Classification: OXIDOREDUCTASE Ligands: NAP, SO4, PEG, GOL, CL |

