Search Count: 59
 |
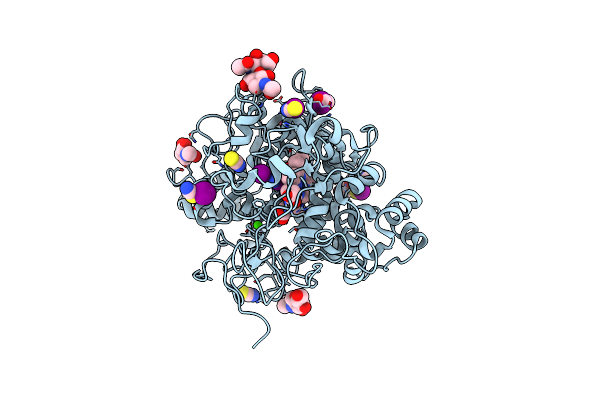
Crystal Structure Of Klebsiella Pneumoniae Enoyl-Acyl Carrier Protein Reductase (Fabi) In Complex With Triclosan
Organism: Klebsiella pneumoniae subsp. pneumoniae 12-3578
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: TCL, NAD, GOL, EDO, SO4 |
 |
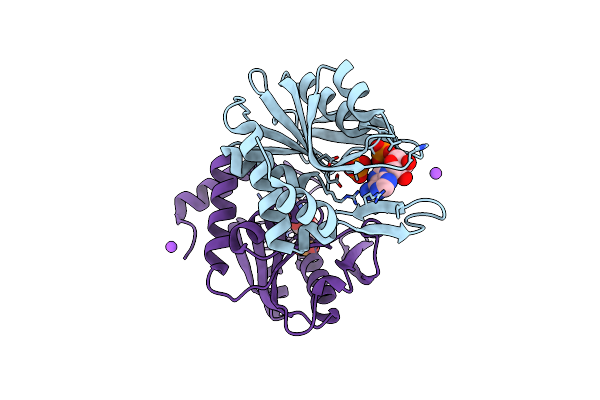
Structure Of The Intermediate Of Lactoperoxidase Formed With Thiocynate And Hydrogen Peroxidase At 1.99 A Resolution.
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: HEM, CA, NAG, IOD, SCN, PEO, GOL |
 |
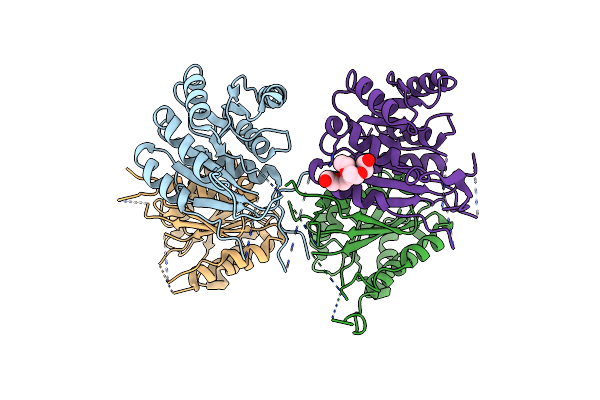
Crystal Structure Of Escherichia Coli Adenine Phosphoribosyltransferase (Aprt) In Complex With Amp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-04-10 Classification: TRANSFERASE Ligands: AMP, CL, PO4, NA |
 |
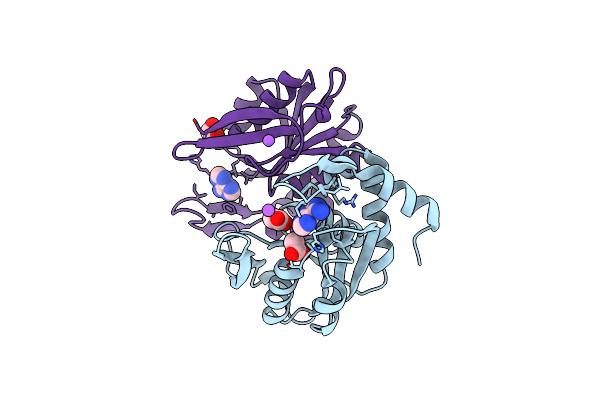
Crystal Structure Of Apo Enoyl-Acyl Carrier Protein Reductase (Fabi) From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae subsp. pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-12-06 Classification: OXIDOREDUCTASE Ligands: P6G |
 |
Crystal Structure Of Escherichia Coli Adenine Phosphoribosyltransferase (Aprt) In Complex With Adenine
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-08-02 Classification: TRANSFERASE Ligands: ADE, ACT, CL, NA, EDO |
 |
Crystal Structure Of Escherichia Coli Adenine Phosphoribosyltransferase (Aprt) In Complex With Phosphate
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-04-12 Classification: TRANSFERASE Ligands: ACT, PO4, CL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2017-06-28 Classification: CHAPERONE Ligands: CL, ACT, FMT, PGE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2015-02-04 Classification: RNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2015-02-04 Classification: RNA BINDING PROTEIN |
 |
Structure Of The Complex Of Type 1 Ribosome Inactivating Protein From Momordica Balsamina With N-Acetylglucosamine At 1.98 Angstrom Resolution Using Crystals Grown In Different Conditions
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-01-14 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Crystal Structure Of Goat Lactoperoxidase In Complex With Octopamine At 2.1 Angstrom Resolution
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-06-18 Classification: OXIDOREDUCTASE Ligands: HEM, NAG, CA, IOD, SCN, OTR, PEG, EDO |
 |
Crystal Structure Of Type 1 Ribosome Inactivating Protein From Momordica Balsamina In Complex With Guanosine Mono Phosphate At 1.75 Angstrom Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2014-05-28 Classification: HYDROLASE Ligands: NAG, 5GP, GOL |
 |
Crystal Structure Of Chitinase D From Serratia Proteamaculans In Complex With N-Acetyl Glucosamine At 1.93 Angstrom Resolution
Organism: Serratia proteamaculans
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2014-04-23 Classification: HYDROLASE Ligands: NAG, ACT, GOL |
 |
Crystal Structure Of Chitinase D From Serratia Proteamaculans In Complex With N-Acetyl Glucosamine, A Hydrolyzed Product Of Hexasaccharide At 1.7 Angstrom Resolution
Organism: Serratia proteamaculans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-04-02 Classification: HYDROLASE Ligands: ACT, NAG, GOL |
 |
Crystal Structure Of The Complex Of Type 1 Ribosome Inactivating Protein From Momordica Balsamina With 5-Fluorouracil At 2.59 A Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2014-01-01 Classification: HYDROLASE Ligands: NAG, GOL, URF |
 |
Crystal Structure Of The Complex Of Ribosome Inactivating Protein From Momordica Balsamina With Peptidoglycan Fragment At 1.78 A Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2013-08-28 Classification: HYDROLASE Ligands: NAG, GOL, NDG, LAC, ALA, GLN |
 |
Crystal Structure Of The Complex Of Ribosome Inactivating Protein From Momordica Balsamina Inhibited By Asymmetric Dimethyl Arginine At 1.70 A Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-08-28 Classification: HYDROLASE Ligands: NAG, DA2 |
 |
Crystal Structure Of Ribosome Inactivating Protein From Momordica Balsamina With Highly Ordered Water Structure In The Substrate Binding Site
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-06-26 Classification: HYDROLASE Ligands: NAG, GOL |
 |
Crystal Structure Of The Complex Of Ribosome Inactivating Protein From Momordica Balsamina With Pyrimidine-2,4(1H,3H)-Dione At 2.57 A Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2013-05-29 Classification: HYDROLASE Ligands: NAG, URA |
 |
Crystal Structure Of Ribosome Inactivating Protein From Momordica Balsamina Complexed With Polyethylene Glycol At 1.90 Angstrom Resolution
Organism: Momordica balsamina
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-05-22 Classification: HYDROLASE Ligands: NAG, PEG, GOL |

