Search Count: 15
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: SOLUTION NMR Release Date: 2024-11-06 Classification: RNA |
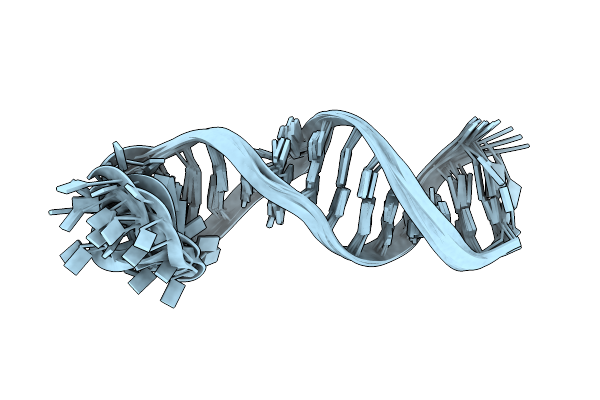 |
Organism: Severe acute respiratory syndrome coronavirus
Method: SOLUTION NMR Release Date: 2024-11-06 Classification: RNA |
 |
Method: ELECTRON MICROSCOPY
Release Date: 2024-10-23 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Procaspase-8 In Complex With Covalent Small Molecule Inhibitor 63-R
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2020-01-29 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 63R |
 |
X-Ray Crystal Structure Of Escherichia Coli Rna Polymerase (Rpob-H526Y) And Ppapp Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2018-01-17 Classification: transferase/transferase inhibitor Ligands: ECJ, MG, ZN |
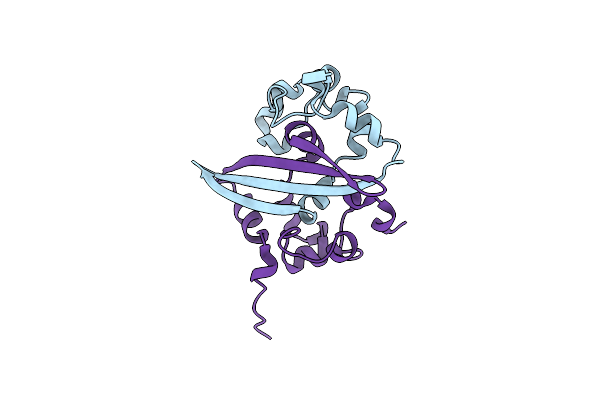 |
Organism: Human coronavirus nl63
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-02-22 Classification: VIRAL PROTEIN |
 |
Organism: Human coronavirus nl63
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-02-22 Classification: RNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Cyprinid herpesvirus 3, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-11-18 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Organism: Cyprinid herpesvirus 3
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-09-11 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
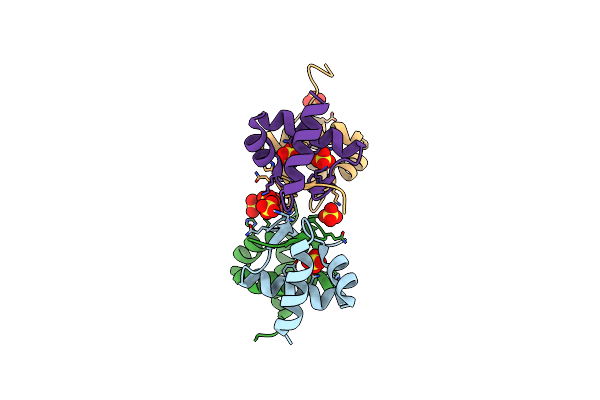
X-Ray Crystal Structure Of Escherichia Coli Sigma70 Holoenzyme In Complex With Guanosine Pentaphosphate (Pppgpp)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2013-04-17 Classification: TRANSCRIPTION, TRANSFERASE Ligands: ZN, 0O2 |
 |
X-Ray Crystal Structure Of Escherichia Coli Sigma70 Holoenzyme In Complex With Guanosine Tetraphosphate (Ppgpp)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2013-04-10 Classification: TRANSCRIPTION, TRANSFERASE Ligands: ZN, G4P |
 |
Apoenzyme Structure Of Homoglutathione Synthetase From Glycine Max In Open Conformation
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-12-22 Classification: LIGASE |
 |
Structure Of Homoglutathione Synthetase From Glycine Max In Open Conformation With Gamma-Glutamyl-Cysteine Bound.
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2009-12-22 Classification: LIGASE Ligands: 3GC |
 |
Structure Of Homoglutathione Synthetase From Glycine Max In Closed Conformation With Homoglutathione, Adp, A Sulfate Ion, And Three Magnesium Ions Bound
Organism: Glycine max
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-12-22 Classification: LIGASE Ligands: ADP, HGS, MG, SO4 |
 |
Crystal Structure Of Homoserine O-Acetyltransferase (Meta) From Bacillus Cereus With Homoserine
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-01-22 Classification: TRANSFERASE Ligands: SO4, HSE |

