Search Count: 24
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cv07-270
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, SO4 |
 |
Crystal Structure Of Sars-Cov-2 Receptor Binding Domain In Complex With Neutralizing Antibody Cv07-250
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2020-10-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-03-06 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The N-Terminal Periplasmic Domain Of Scsb From Proteus Mirabilis
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2018-03-07 Classification: OXIDOREDUCTASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2017-08-02 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2017-07-26 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-06-08 Classification: ISOMERASE |
 |
Organism: Acinetobacter baumannii aye, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-06-18 Classification: TRANSLATION/OXIDOREDUCTASE Ligands: MG, GDP, GOL |
 |
Crystal Structure Of The Disulfide Oxidoreductase Dsba From Proteus Mirabilis
Organism: Proteus mirabilis hi4320
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: MLI |
 |
Crystal Structure Of The Disulfide Oxidoreductase Dsba (S30Xxc33) Active Site Mutant From Proteus Mirabilis
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE Ligands: SCN |
 |
Complex Structure Of Proteus Mirablis Dsba (C30S) With A Non-Covalently Bound Peptide Pwatcds
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-05-28 Classification: OXIDOREDUCTASE/PEPTIDE Ligands: SCN |
 |
Organism: Klebsiella pneumoniae
Method: SOLUTION NMR Release Date: 2013-12-11 Classification: OXIDOREDUCTASE |
 |
Disulfide Isomerase (Dsbp) From Multidrug Resistance Inca/C Transferable Plasmid In Oxidized State (P212121 Space Group)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-12-11 Classification: ISOMERASE |
 |
Disulfide Isomerase From Multidrug Resistance Inca/C Conjugative Plasmid In Reduced State
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-12-11 Classification: ISOMERASE |
 |
Disulfide Isomerase From Multidrug Resistance Inca/C Related Integrative And Conjugative Elements In Oxidized State (P21 Space Group)
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2013-12-11 Classification: ISOMERASE Ligands: BU2 |
 |
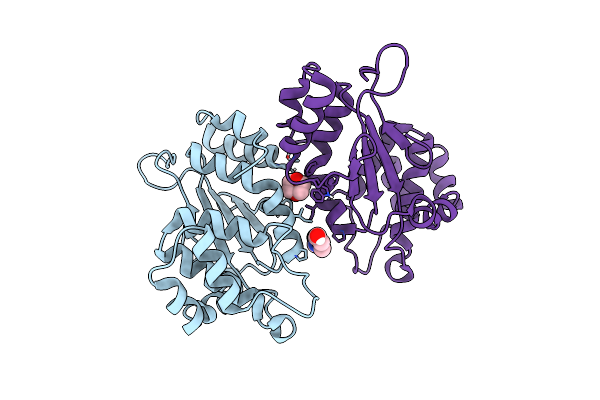
Crystal Structure Of Disulfide Oxidoreductase From Klebsiella Pneumoniae In Reduced State
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2013-11-27 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Disulfide Oxidoreductase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-10-02 Classification: OXIDOREDUCTASE Ligands: EPE, DIO |
 |
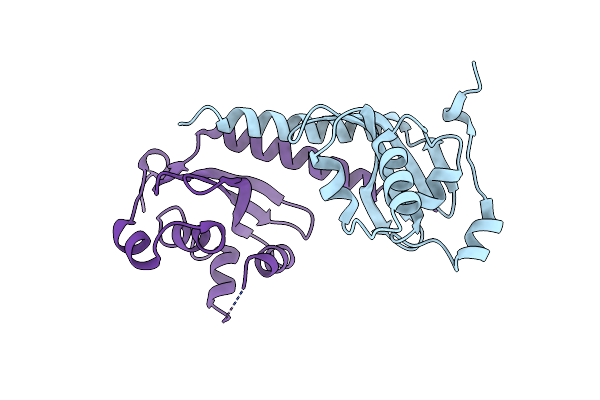
Crystal Structure Of A Periplasmic Thioredoxin-Like Protein From Salmonella Enterica Serovar Typhimurium
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2013-07-17 Classification: ISOMERASE |
 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-10-31 Classification: TRANSCRIPTION |

