Search Count: 154
 |
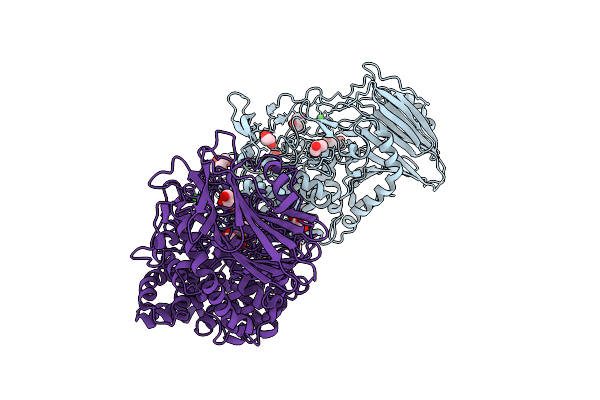
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: GOL, CA, TLA |
 |
Crystal Structure Of Tegh116 From Thermosynechococcus Elongatus With Glucose
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: HYDROLASE Ligands: BGC, GOL, CA, TLA |
 |
X-Ray Structure Of Cytochrome P450 Olet From Lacicoccus Alkaliphilus In Complex With Icosanoic Acid
Organism: Lacicoccus alkaliphilus dsm 16010
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: HEM, DCR, GOL |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Manganese (Mn) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Iron (Fe) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of A Cupin Protein (Tm1459) In Zinc (Zn) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of A Cupin Protein (Tm1459, C106V Mutant) In Iron (Fe) Substituted Form
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
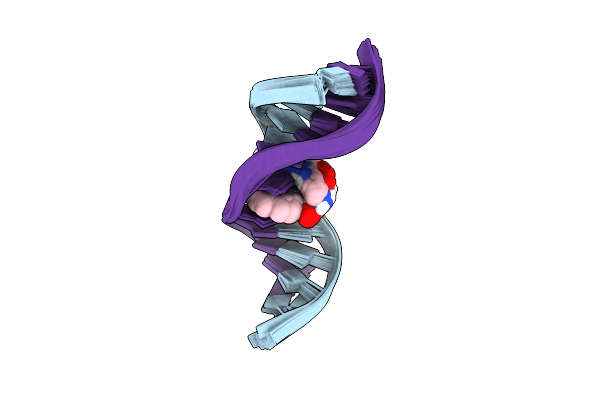
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
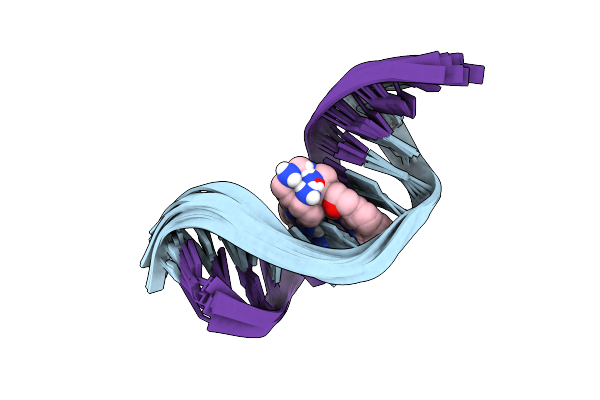
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1R |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-04-30 Classification: DNA Ligands: A1L1S |
 |
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MOTOR PROTEIN |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Reduced Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
Neutron Structure Of Ferredoxin-Nadp+ Reductase From Maize Root -Oxidized Form
Organism: Zea mays
Method: NEUTRON DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, GOL, FAD |
 |
High Resolution Structure Of Ferredoxin-Nadp+ Reductase From Maize Root - Reduced Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: MES, FDA |
 |
R115A Mutant Of Ferredoxin-Nadp+ Reductase From Maize Root - Oxidized Form, Low X-Ray Dose
Organism: Zea mays
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, MES |
 |
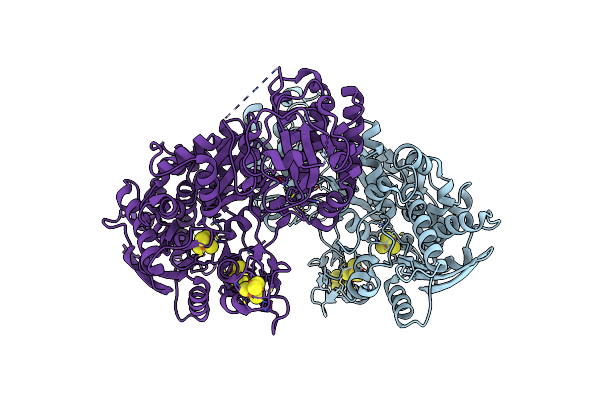
Organism: Clostridium beijerinckii
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: OXIDOREDUCTASE Ligands: ZN, SF4, 402 |
 |
Crystal Structure Of Apo-[Fefe]-Hydrogenase Cba5H From Clostridium Beijerinckii
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: SF4, ZN, CL |
 |
Crystal Structure Of [Fefe]-Hydrogenase Cba5H From Clostridium Beijerinckii In Hinact State
Organism: Clostridium beijerinckii
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: 402, SF4, ZN, CL |
 |
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2024-11-20 Classification: PHOTOSYNTHESIS Ligands: CHL, CLA, A1LXP, NEX, LHG, XAT |

