Search Count: 40
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FDA, NAD |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Organism: Sus scrofa
Method: NEUTRON DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Organism: Sus scrofa
Method: NEUTRON DIFFRACTION Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION Resolution:0.86 Å Release Date: 2024-02-14 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
 |
Organism: Alcaligenes faecalis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.50 Å, 1.90 Å Release Date: 2024-02-14 Classification: ELECTRON TRANSPORT Ligands: CU, SO4, DOD |
 |
Crystal Structure Of A De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 (K4T) - Pt Derivative
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-09-20 Classification: DE NOVO PROTEIN Ligands: PT, CL, ACT, PC4 |
 |
Crystal Structure Of A De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 (K4T)
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-20 Classification: DE NOVO PROTEIN Ligands: ACT |
 |
Crystal Structure Of A De Novo Enzyme, Ferric Enterobactin Esterase Syn-F4 (K4T) At 2.0 Angstrom Resolution
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-09-20 Classification: DE NOVO PROTEIN Ligands: ACT |
 |
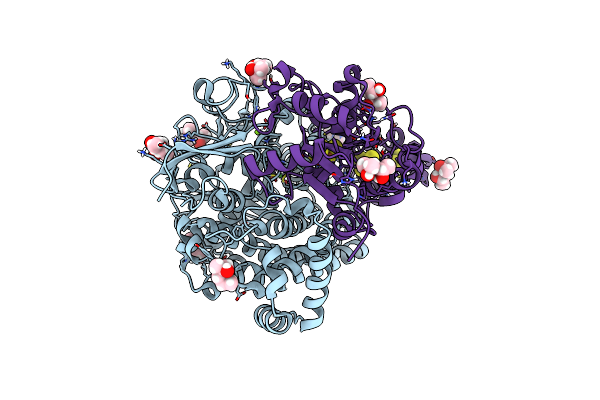
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Globiformis Anaerobically Reduced By Phenylethylamine At Pd 9.0
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.09 Å, 1.67 Å Release Date: 2023-09-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, PEA |
 |
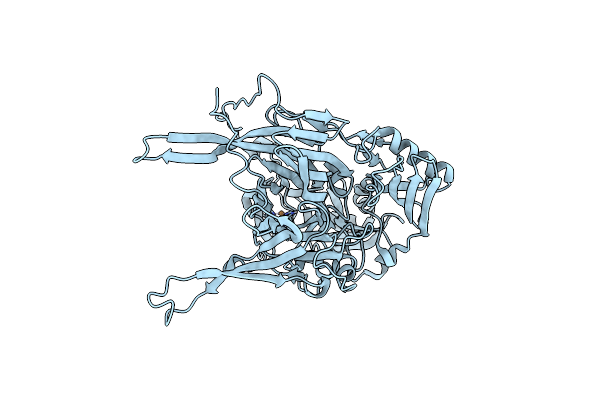
Neutron Structure Of [Nife]-Hydrogenase From D. Vulgaris Miyazaki F In Its Oxidized State
Organism: Desulfovibrio vulgaris str. 'miyazaki f'
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.04 Å, 2.20 Å Release Date: 2023-09-13 Classification: OXIDOREDUCTASE Ligands: NFU, MPD, OH, MG, CL, SF4, F3S |
 |
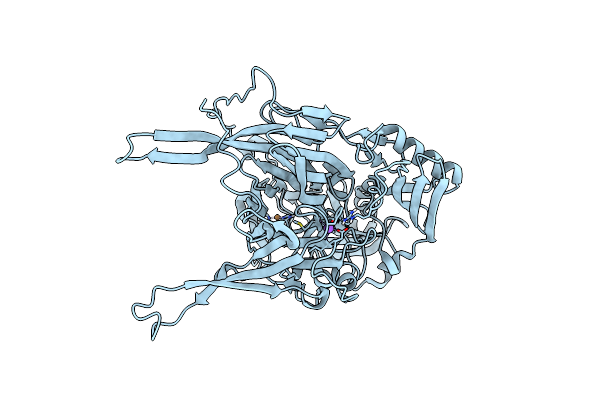
Crystallographic Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4 Determined By Only Neutron Diffraction Data.
Organism: Arthrobacter globiformis
Method: NEUTRON DIFFRACTION Resolution:1.72 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: CU, DOD |
 |
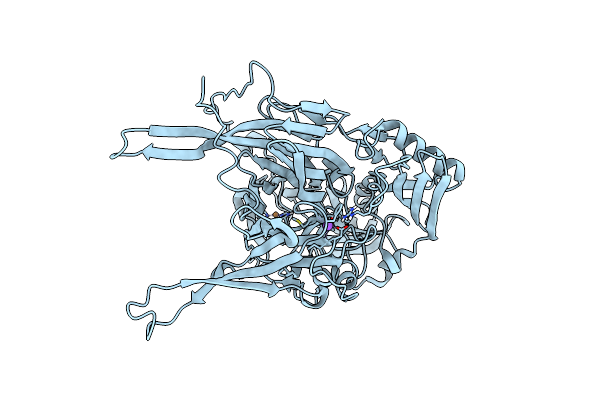
Crystallographic Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4 Determined By Both X-Ray And Neutron Diffraction Data At 1.72 Angstrom Resolution.
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.7200 Å, 1.7200 Å Release Date: 2022-04-20 Classification: OXIDOREDUCTASE Ligands: CU, NA, DOD |
 |
Neutron Structure Of Copper Amine Oxidase From Arthrobacter Glibiformis At Pd 7.4
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.1400 Å, 1.7200 Å Release Date: 2020-04-29 Classification: OXIDOREDUCTASE Ligands: CU, NA |
 |
Organism: Halorhodospira halophila
Method: NEUTRON DIFFRACTION Resolution:1.49 Å Release Date: 2017-08-30 Classification: SIGNALING PROTEIN Ligands: HC4, DOD |
 |
High Resolution Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2013-09-11 Classification: OXIDOREDUCTASE Ligands: CU, NA, EDO, PEG, PGE, PG4, 1PE, OXY |
 |
Crystal Structure Of Copper Amine Oxidase From Arthrobacter Globiformis In N2 Condition
Organism: Arthrobacter globiformis
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-09-11 Classification: OXIDOREDUCTASE Ligands: CU, NA, PGE, PG4, 1PE, EDO, PEG, HDZ |
 |
X/N Joint Refinement Of Achromobacter Lyticus Protease I Free Form At Pd8.0
Organism: Achromobacter lyticus
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.895 Å, 1.979 Å Release Date: 2013-09-11 Classification: HYDROLASE Ligands: DOD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2013-09-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG |
 |
Organism: Unidentified, Homo sapiens
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Resolution:1.602 Å, 2.752 Å Release Date: 2013-09-04 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG, DOD |

