Search Count: 36
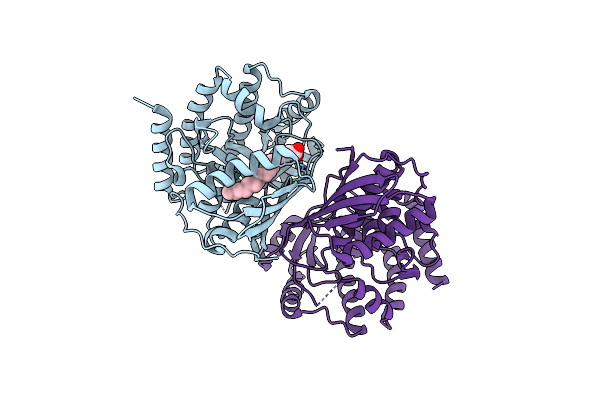 |
Crystal Structure Of Mevalonate 3,5-Bisphosphate Decarboxylase From Picrophilus Torridus
Organism: Picrophilus torridus dsm 9790
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2021-12-22 Classification: LYASE Ligands: OLA |
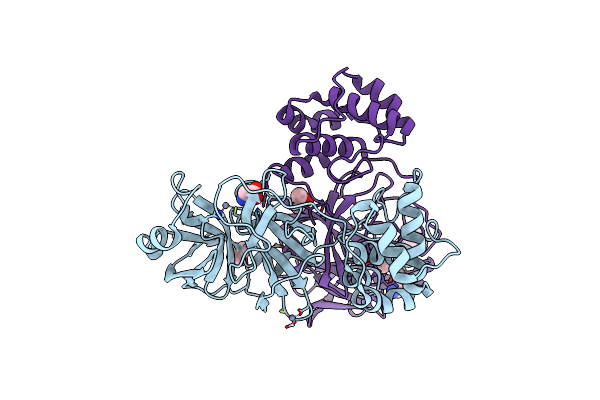 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: ZN, DMS, GLY, CL, BTB |
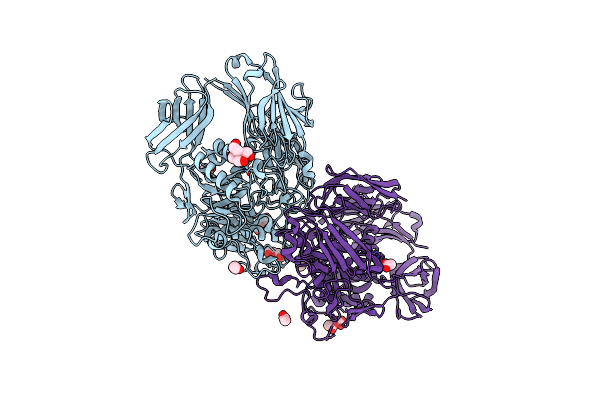 |
Crystal Structure Of A Cold-Adapted Dimeric Beta-D-Galactosidase From Paracoccus Sp. 32D Strain
Organism: Paracoccus sp. 32d
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-09-14 Classification: HYDROLASE Ligands: ACT, BTB, PEG |
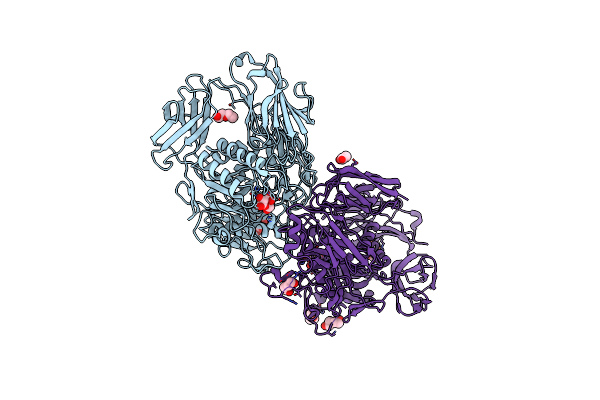 |
Crystal Structure Of A Cold-Adapted Dimeric Beta-D-Galactosidase From Paracoccus Sp. 32D Strain In Complex With Galactose
Organism: Paracoccus sp. 32d
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2016-09-14 Classification: HYDROLASE Ligands: GAL, GLC, PEG, CL, ACT |
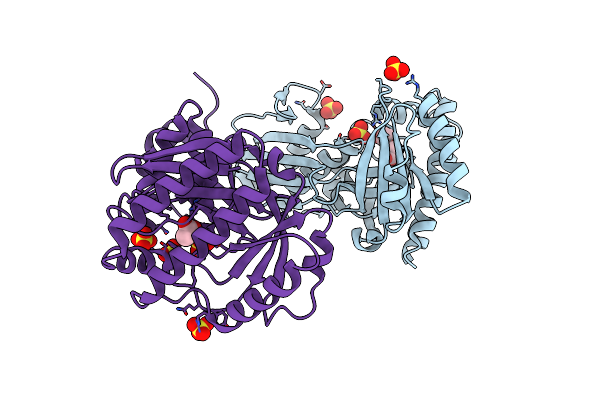 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Apo Form)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: SO4, ACT |
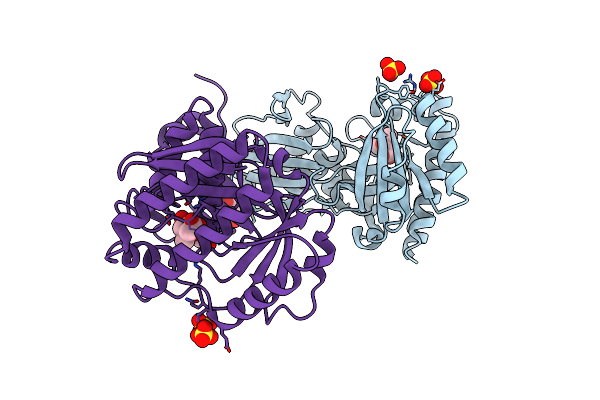 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Mevalonate Bound)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: MEV, SO4, ACT, GOL |
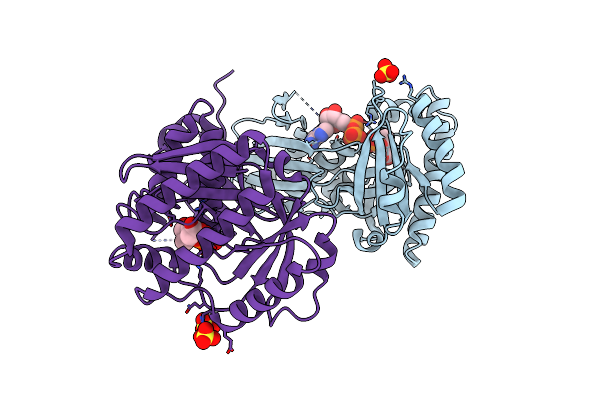 |
Crystal Structure Of Mevalonate-3-Kinase From Thermoplasma Acidophilum (Mevalonate 3-Phosphate/Adp Bound)
Organism: Thermoplasma acidophilum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-12-10 Classification: TRANSFERASE Ligands: ADP, 3S4, SO4 |
 |
Crystal Structure Of Methylthioadenosine Phosphorylase Sourced From An Antarctic Soil Metagenomic Library
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-02-20 Classification: TRANSFERASE |
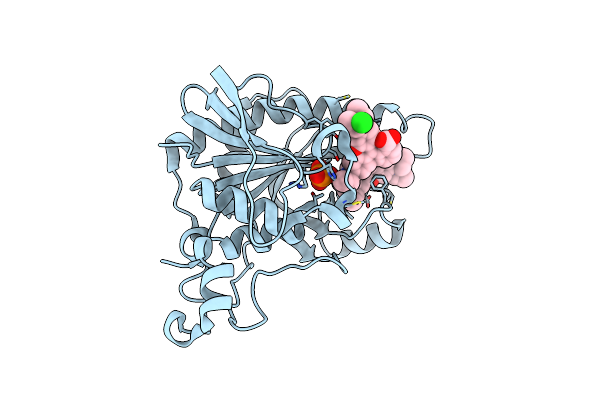 |
Crystal Structure Of Methylthioadenosine Phosphorylase In Complex With Rhodamine B
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-02-20 Classification: TRANSFERASE Ligands: RHB, PO4, CL |
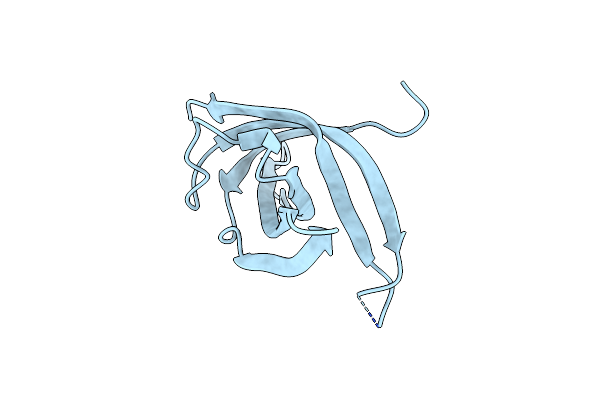 |
Dimeric Structure Of The N-Terminal Domain Of Prib Protein From Thermoanaerobacter Tencongensis Solved Ab Initio
Organism: Thermoanaerobacter tengcongensis
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2012-09-19 Classification: DNA BINDING PROTEIN |
 |
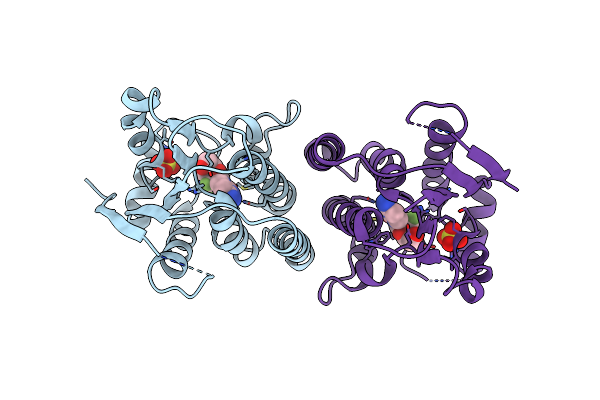
Crystal Structure Of Psychrotrophic Esterase Esta From Pseudoalteromonas Sp. 643A Inhibited By Monoethylphosphonate
Organism: Pseudoalteromonas sp.
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2009-06-30 Classification: HYDROLASE |
 |
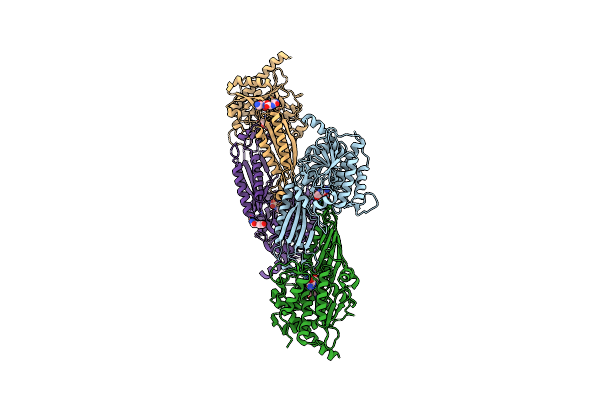
Drosophila Melanogaster Deoxyribonucleoside Kinase Successfully Activates Gemcitabine In Transduced Cancer Cell Lines
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-03-24 Classification: TRANSFERASE Ligands: GEO, SO4 |
 |
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With A Gly-Gly Peptide
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2008-05-13 Classification: HYDROLASE Ligands: GLY, ZN |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2008-03-25 Classification: TRANSCRIPTION |
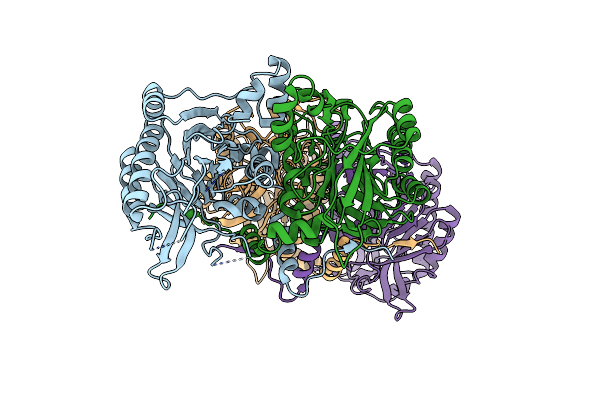 |
Crystal Structure Of A Pyrimidine Degrading Enzyme From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-03-25 Classification: HYDROLASE |
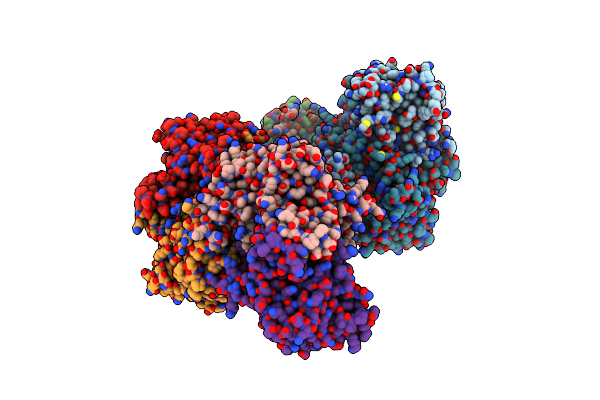 |
Crystal Structure Of A Pyrimidine Degrading Enzyme From Drosophila Melanogaster
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-03-25 Classification: HYDROLASE |
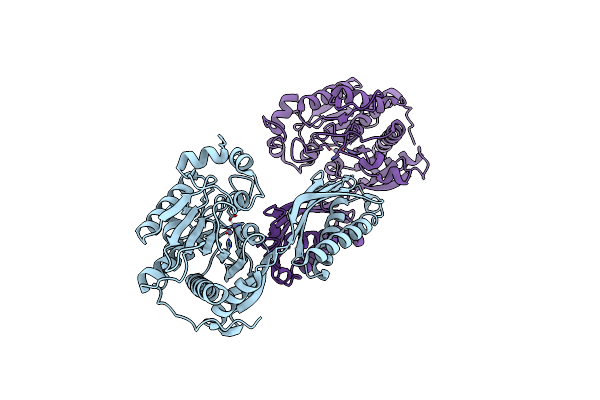 |
Crystal Structure Of Mutant E159A Of Beta-Alanine Synthase From Saccharomyces Kluyveri
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN |
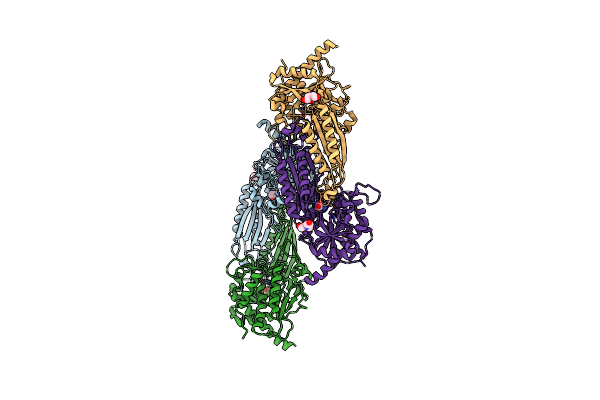 |
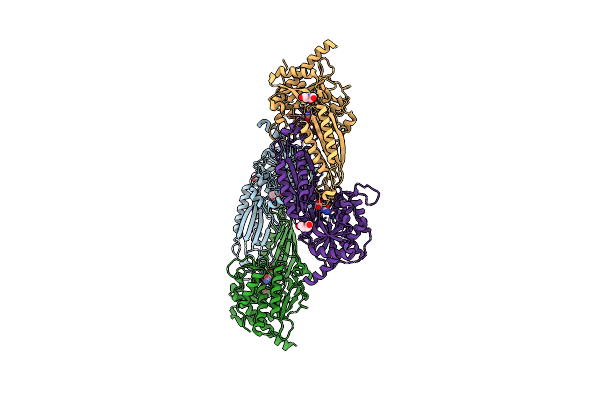
Crystal Structure Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With The Product Beta-Alanine
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, BAL, BCN |
 |
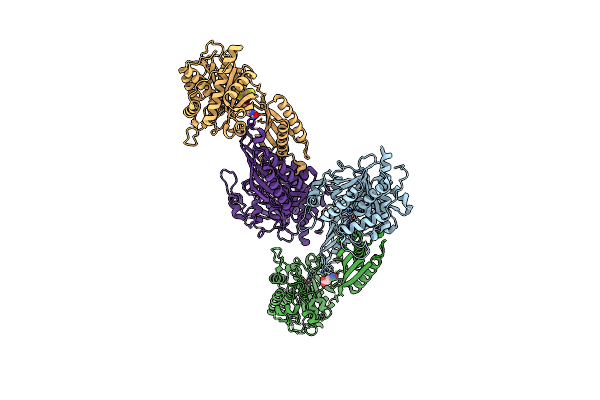
Crystal Structure Of Mutant E159A Of Beta-Alanine Synthase From Saccharomyces Kluyveri In Complex With Its Substrate N-Carbamyl-Beta- Alanine
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, URP, BCN |
 |
Crystal Structure Of Mutant R322A Of Beta-Alanine Synthase From Saccharomyces Kluyveri
Organism: Saccharomyces kluyveri
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2007-10-02 Classification: HYDROLASE Ligands: ZN, URP, DTT |

