Search Count: 37
 |
Organism: Aspergillus oryzae
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SO4 |
 |
Crystal Structure Of An Exoribonuclease-Resistant Rna From A Tombusvirus-Like Associated Rna
Organism: Beet western yellows st9 associated virus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: RNA Ligands: IRI, MG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of A Sur1/Kir6.2-Q52R Atp-Sensitive Potassium Channel In The Presence Of Pip2 In The Open Conformation
Organism: Rattus norvegicus, Mesocricetus auratus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: PT5, K, PEF, P5S, NAG |
 |
Cryo-Em Structure Of A Sur1/Kir6.2-Q52R Atp-Sensitive Potassium Channel In The Presence Of Pip2 In The Open Conformation
Organism: Rattus norvegicus, Mesocricetus auratus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: PT5, K, PEF, P5S, NAG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2023-03-29 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Human Rxralpha Ligand Binding Domain In Complex With 2,4-Di-Tert-Butylphenol And A Coactivator Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-03-02 Classification: GENE REGULATION Ligands: UGW, FMT |
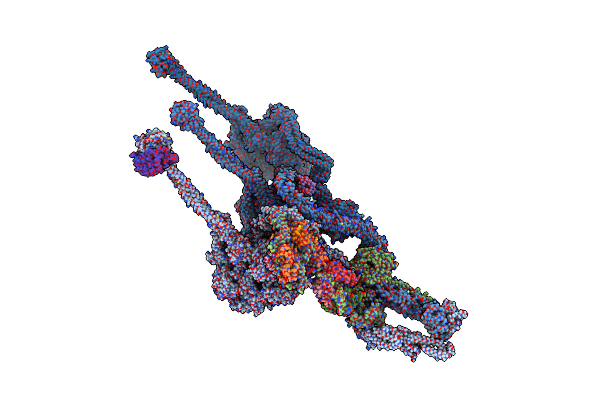 |
Structure Of Outer-Arm Dyneins Bound To Microtubule With Microtubule Binding State 1(Mtbs-1)
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Resolution:4.00 Å Release Date: 2021-09-29 Classification: STRUCTURAL PROTEIN Ligands: ADP, ATP, MG |
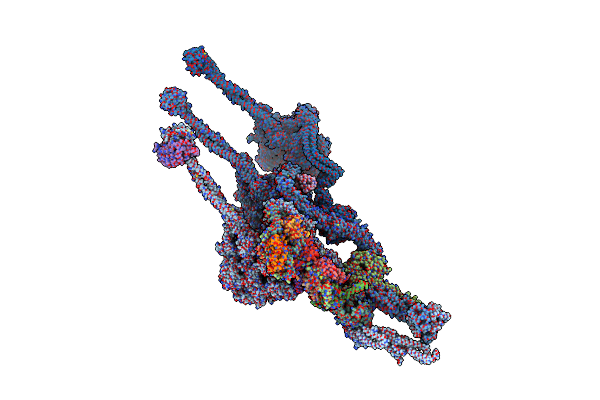 |
Structure Of Outer-Arm Dynein Bound To Microtubule Doublet In Microtubule Binding State 2 (Mtbs-2)
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: MOTOR PROTEIN Ligands: ADP, ATP, MG |
 |
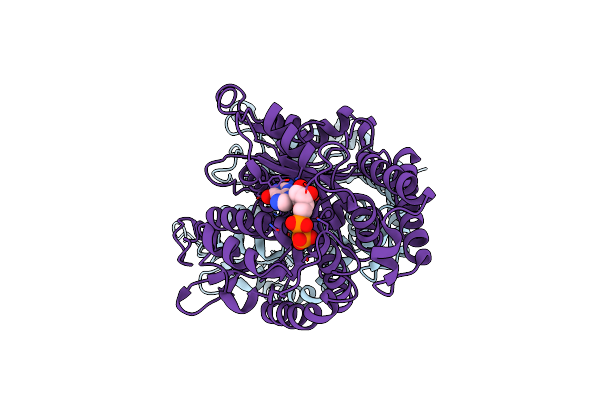
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: MOTOR PROTEIN Ligands: ADP, ATP, MG |
 |
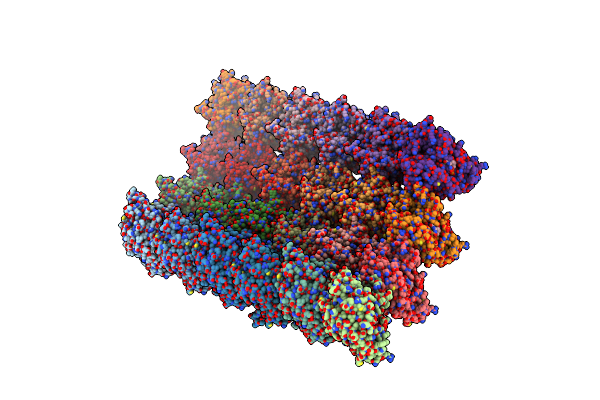
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Release Date: 2021-09-29 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP |
 |
Organism: Tetrahymena thermophila
Method: ELECTRON MICROSCOPY Resolution:4.50 Å Release Date: 2021-09-29 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP |
 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-07-28 Classification: SIGNALING PROTEIN Ligands: GNP, MG |
 |
Crystal Structure Of The N-Terminal Fragment (Residue 1-206) Of Lona Protease From Meiothermus Taiwanensis
Organism: Meiothermus taiwanensis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-05-26 Classification: HYDROLASE |
 |
Crystal Structure Of The N-Terminal Fragment (Residue 1-291) Of Lona Protease From Meiothermus Taiwanensis
Organism: Meiothermus taiwanensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-05-26 Classification: HYDROLASE Ligands: SO4 |
 |
Structure Of Bifidobacterium Dentium Beta-Glucuronidase Complexed With C6-Hexyl Uronic Isofagomine
Organism: Bifidobacterium dentium (strain atcc 27534 / dsm 20436 / jcm 1195 / bd1)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-01-27 Classification: CARBOHYDRATE Ligands: E8X |
 |
Organism: Bifidobacterium dentium (strain atcc 27534 / dsm 20436 / jcm 1195 / bd1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-01-27 Classification: HYDROLASE |
 |
Structure Of Bifidobacterium Dentium Beta-Glucuronidase Complexed With Uronic Isofagomine
Organism: Bifidobacterium dentium (strain atcc 27534 / dsm 20436 / jcm 1195 / bd1)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: SJ5 |
 |
Structure Of Bifidobacterium Dentium Beta-Glucuronidase Complexed With C6-Nonyl Uronic Isofagomine
Organism: Bifidobacterium dentium (strain atcc 27534 / dsm 20436 / jcm 1195 / bd1)
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2021-01-27 Classification: HYDROLASE Ligands: E9O |

