Search Count: 32
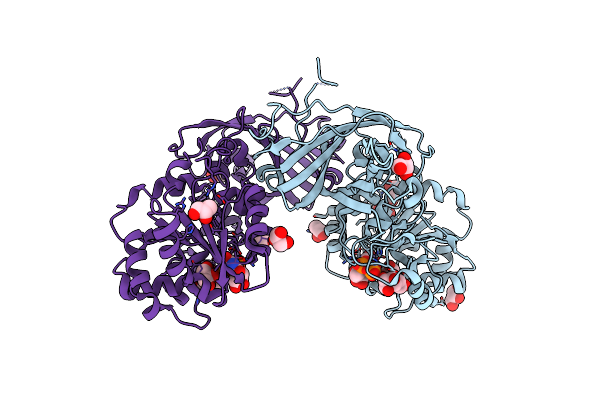 |
Crystal Structure Of Aspergillus Fumigatus Galactofuranosylransferase (Afgfsa) In Complex With Udp And Galactofuranose
Organism: Aspergillus fumigatus cea10
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2024-10-30 Classification: TRANSFERASE Ligands: MN, UDP, GOL, GZL |
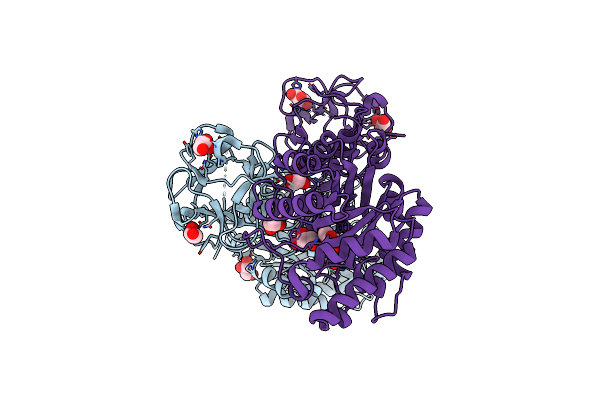 |
Crystal Structure Of Covalent Glycosyl-Enzyme Intermediate Of Xylanase Mutant (T82A, N127S, And E128H) From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: GOL |
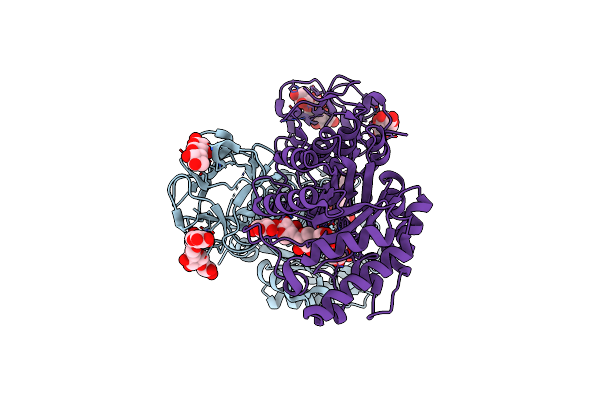 |
Crystal Structure Of Michaelis Complex Of Xylanase Mutant (T82A, N127S, And E128H) From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-09 Classification: HYDROLASE |
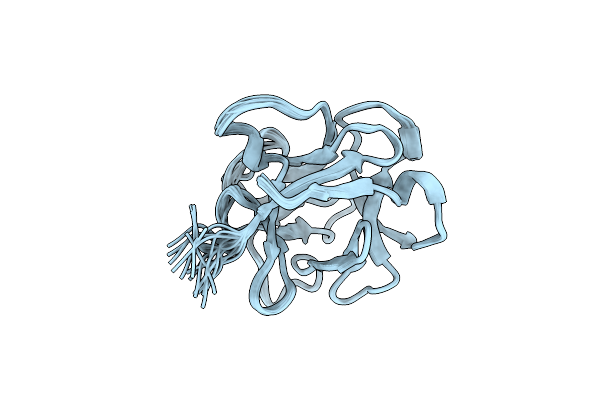 |
Organism: Lumbricus terrestris
Method: SOLUTION NMR Release Date: 2013-04-17 Classification: SUGAR BINDING PROTEIN |
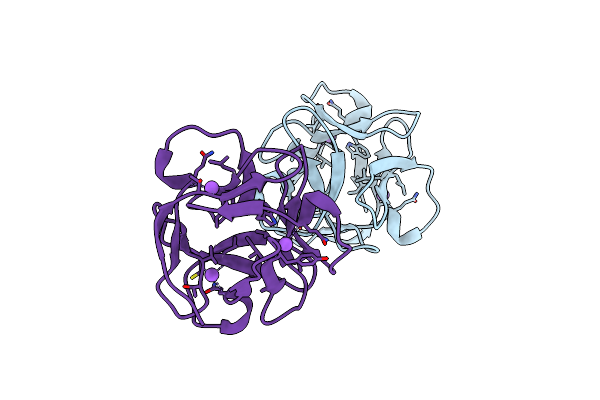 |
Organism: Actinomycete
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2009-08-25 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of The Earthworm R-Type Lectin C-Half In Complex With Lactose
Organism: Lumbricus terrestris
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2008-09-02 Classification: SUGAR BINDING PROTEIN Ligands: PO4, IMD |
 |
Crystal Structure Of The Earthworm R-Type Lectin C-Half In Complex With Galnac
Organism: Lumbricus terrestris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-09-02 Classification: SUGAR BINDING PROTEIN Ligands: NGA, CD, PO4, IMD |
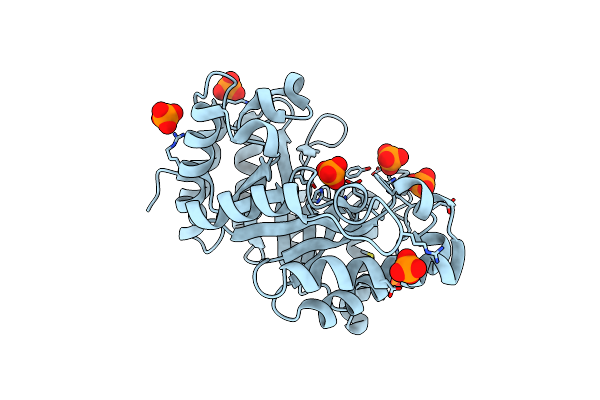 |
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-03-06 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2007-03-06 Classification: HYDROLASE Ligands: PO4 |
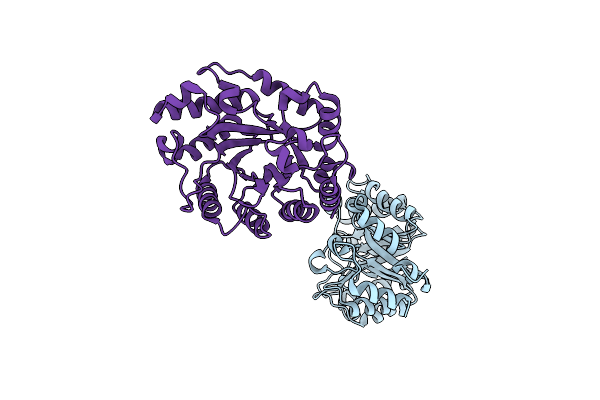 |
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2007-03-06 Classification: HYDROLASE |
 |
Organism: Lumbricus terrestris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-02-06 Classification: SUGAR BINDING PROTEIN Ligands: SO4, PGE |
 |
Crystal Structure Of The Earthworm Lectin C-Terminal Domain Mutant In Complex With Lactose
Organism: Lumbricus terrestris
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2007-02-06 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of The Earthworm Lectin C-Terminal Domain Mutant In Complex With 6'-Sialyllactose
Organism: Lumbricus terrestris
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-02-06 Classification: SUGAR BINDING PROTEIN Ligands: SO4 |
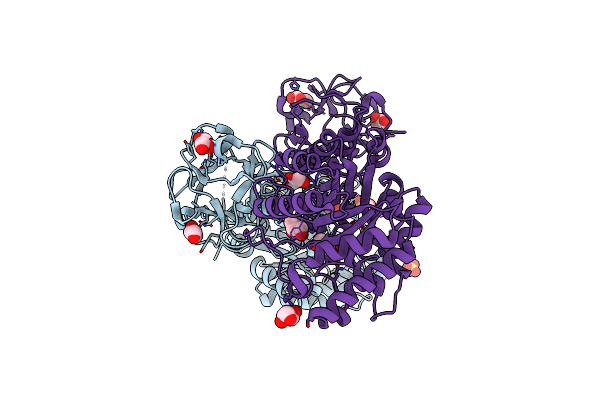 |
Crystal Structure Of Catalytic-Site Mutant Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2006-10-10 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of Michaelis Complex Of Catalytic-Site Mutant Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-10-10 Classification: HYDROLASE Ligands: NPO, GOL |
 |
Crystal Structure Of Covalent Glycosyl-Enzyme Intermediate Of Catalytic-Site Mutant Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-10-10 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of Ep Complex Of Catalytic-Site Mutant Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2006-10-10 Classification: HYDROLASE Ligands: AZI, GOL |
 |
Crystal Structure Of Es Complex Of Catalytic-Site Mutant Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-10-10 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Organism: Aspergillus kawachii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-09-19 Classification: HYDROLASE |
 |
Crystal Structure Of Arabinofuranosidase Complexed With Arabinofuranosyl-Alpha-1,2-Xylobiose
Organism: Aspergillus kawachii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-09-19 Classification: HYDROLASE |

