Search Count: 41
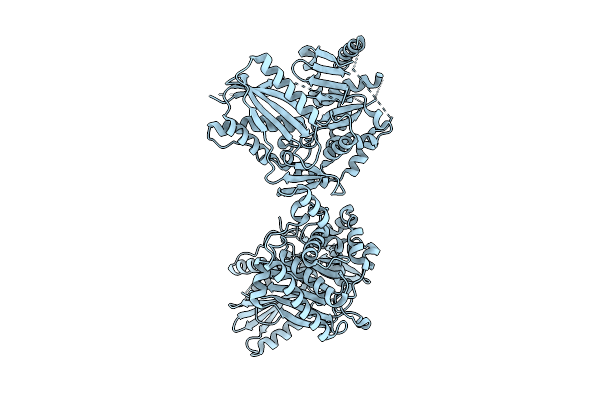 |
Crystal Structure Of The B. Licheniformis Bacitacin Sythetase 3 Cis-E-Com-C Domains
Organism: Bacillus licheniformis
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: PEPTIDE BINDING PROTEIN |
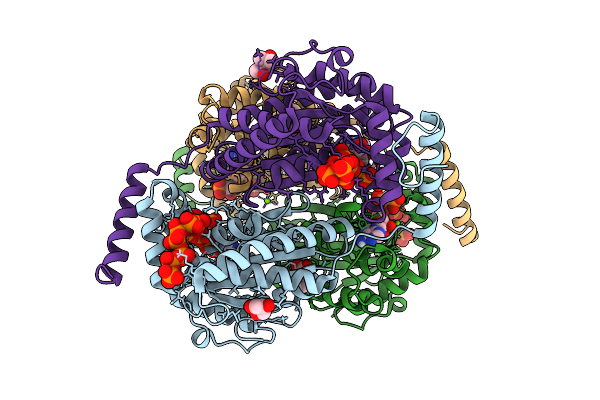 |
Crystal Structure Of Ppk2 Class Iii From Erysipelotrichaceae Bacterium In Complex With Appch2P And Polyphosphate
Organism: Erysipelotrichaceae bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: ACP, 6YY, GOL, BEZ, MG, MPD, A1I4D |
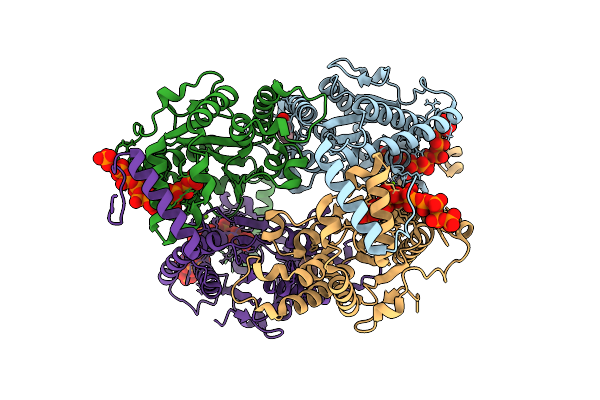 |
Crystal Structure Of Ppk2 Class Iii From Erysipelotrichaceae Bacterium In Complex With Polyphosphate
Organism: Erysipelotrichaceae bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: TRANSFERASE Ligands: 6YY, BEZ, PO4, MG |
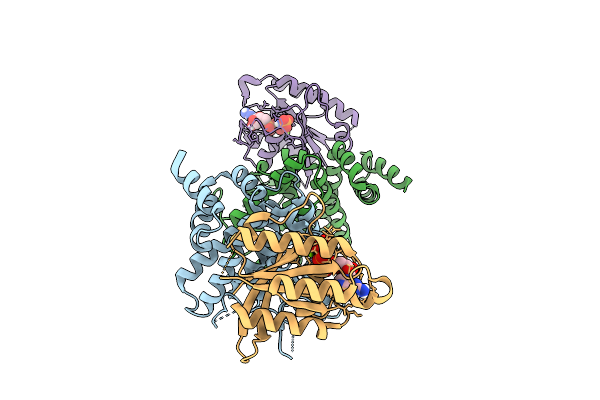 |
Crystal Structure Of The Human Ralgapa2 N-Terminal Domain With Human Kappab-Ras1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: SIGNALING PROTEIN Ligands: GNP, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
Crystal Structure Of The Fused, Cysteine-Less Aes123 Polb1 Split Intein (With S1A, N159A Mutations)
Organism: Aeromonas phage aes123
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2025-05-07 Classification: SPLICING |
 |
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-07-26 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, MLI, MG |
 |
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Release Date: 2023-05-17 Classification: ENDOCYTOSIS |
 |
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-05-17 Classification: SPLICING Ligands: IOD |
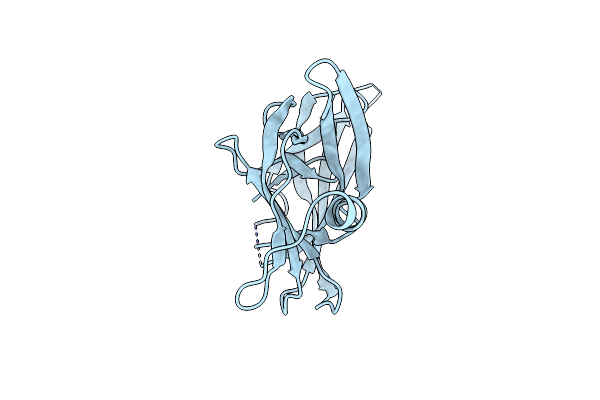 |
Crystal Structure Of The Polb16_Oarg Intein Variant S1A, N183A, C111A, C165A
Organism: Metagenome
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-05-17 Classification: SPLICING |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2023-05-10 Classification: Hydrolase, Isomerase Ligands: 12P, ZN, CL |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2023-05-10 Classification: Hydrolase, Isomerase Ligands: ZN |
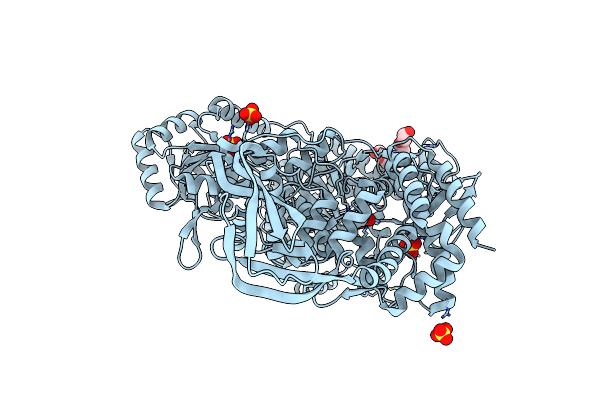 |
Crystal Structure Of T. Maritima Reverse Gyrase With A Minimal Latch, Hexagonal Form
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2023-04-26 Classification: HYDROLASE Ligands: P6G, ZN, SO4, CL |
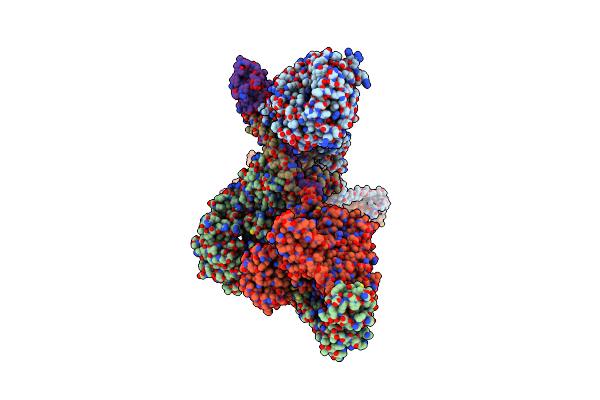 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-09-28 Classification: CYTOSOLIC PROTEIN |
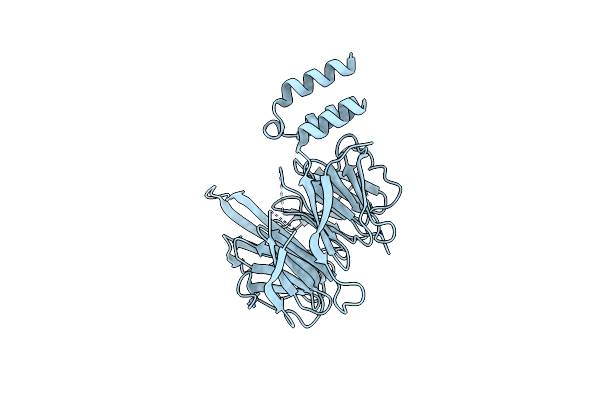 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2022-09-21 Classification: TRANSPORT PROTEIN |
 |
Organism: Chaetomium thermophilum
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: ENDOCYTOSIS |
 |
Chlamydomonas Reinhardtii Nadph Dependent Thioredoxin Reductase 1 Domain Cs Mutant
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2022-01-19 Classification: OXIDOREDUCTASE Ligands: FAD, P4G |
 |
Crystal Structure Of Chlamydomonas Reinhardtii Nadph Dependent Thioredoxin Reductase 1 Domain
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Crystal Structure Of Apo Form L147A/I351A Variant Of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2021-11-17 Classification: TRANSFERASE |
 |
Crystal Structure Of Apo Form Of S-Adenosylmethionine Synthetase From Methanocaldococcus Jannaschii
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2021-11-17 Classification: TRANSFERASE |

