Search Count: 9
 |
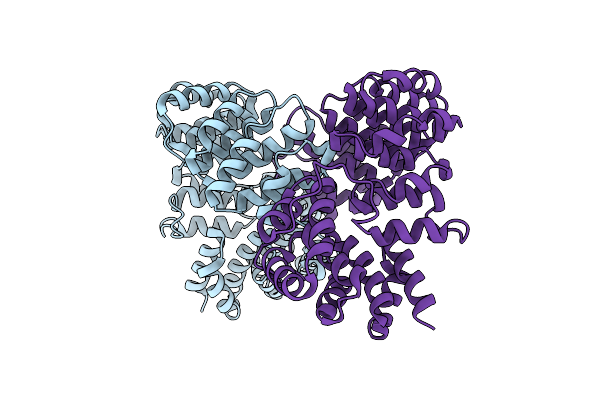
Crystal Structure Of The Histidine Kinase Domain Of Bacteriophytochrome Rpbphp2
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:3.19 Å Release Date: 2023-11-22 Classification: SIGNALING PROTEIN, TRANSFERASE Ligands: ATP, MG |
 |
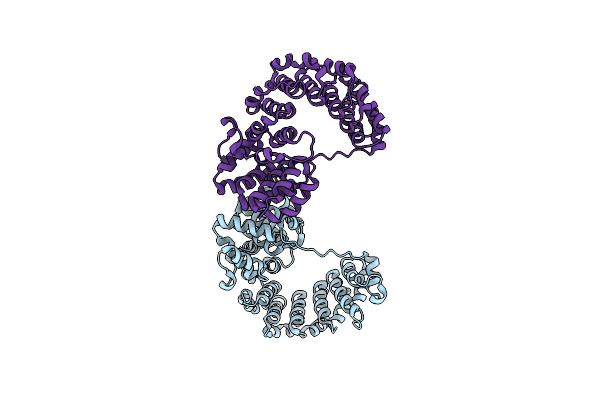
Crystal Structure Of A Single-Chain E/F Type Bilin Lyase-Isomerase Mpeq In Space Group C2221
Organism: Synechococcus sp. a15-62
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2022-08-24 Classification: LYASE |
 |
Organism: Synechococcus sp. p1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-02-23 Classification: LYASE |
 |
Complete Lov Domain From The Lov-Hk Sensory Protein From Brucella Abortus (Mutant C69S, Construct 15-155)
Organism: Brucella melitensis biotype 1 (strain 16m / atcc 23456 / nctc 10094)
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2020-12-30 Classification: TRANSFERASE Ligands: FMN |
 |
Lov-Pas Construct From The Lov-Hk Sensory Protein From Brucella Abortus (Dark-Adapted, Construct 15-273)
Organism: Brucella melitensis biotype 1 (strain 16m / atcc 23456 / nctc 10094)
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2020-12-30 Classification: TRANSFERASE Ligands: FMN |
 |
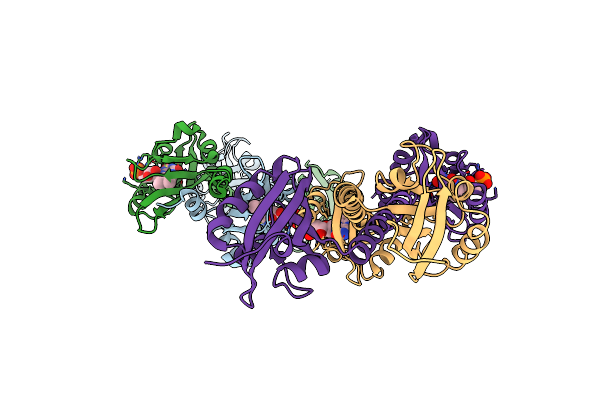
Full Length Lov-Pas-Hk Construct From The Lov-Hk Sensory Protein From Brucella Abortus (Light-Adapted, Construct 15-489)
Organism: Brucella melitensis biotype 1 (strain 16m / atcc 23456 / nctc 10094)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2020-12-30 Classification: TRANSFERASE Ligands: CL, FMN, MG, ACP |
 |
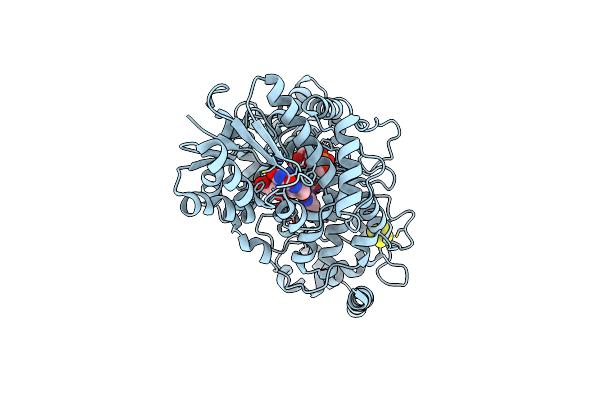
A Blue Light Illuminated Lov-Pas Construct From The Lov-Hk Sensory Protein From Brucella Abortus (Construct 15-273)
Organism: Brucella abortus (strain 2308)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-07-15 Classification: SIGNALING PROTEIN Ligands: FMN |
 |
Crystal Structure Of Bacterial (6-4) Photolyase Phrb From In Situ Serial Laue Diffraction
Organism: Rhizobium radiobacter
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-07-18 Classification: LYASE Ligands: FAD, DLZ, SF4 |
 |
Crystal Structure Of Plant Uvb Photoreceptor Uvr8 From In Situ Serial Laue Diffraction
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-07-18 Classification: GENE REGULATION |

