Search Count: 402
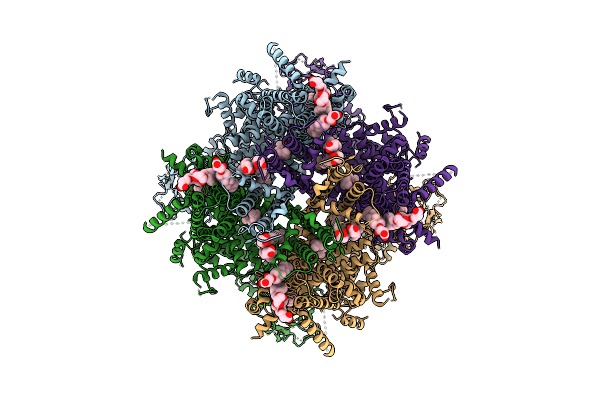 |
Cryo-Em Structure Of Rabbit Trpm3 In Apo Resting State At 37 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
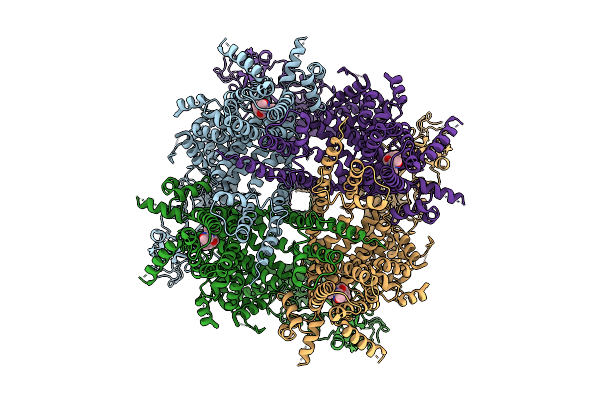 |
Cryo-Em Structure Of Rabbit Trpm3 In Complex With Cim0216 In Resting State At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: A1AIB |
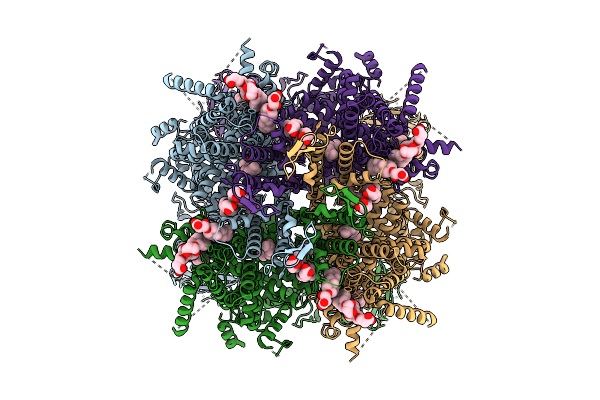 |
Cryo-Em Structure Of Rabbit Trpm3 In Apo Activated State At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
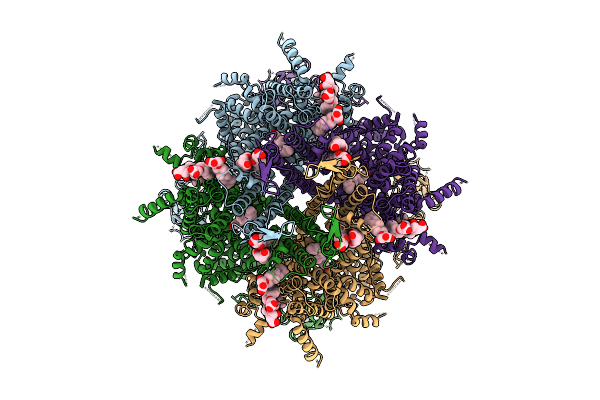 |
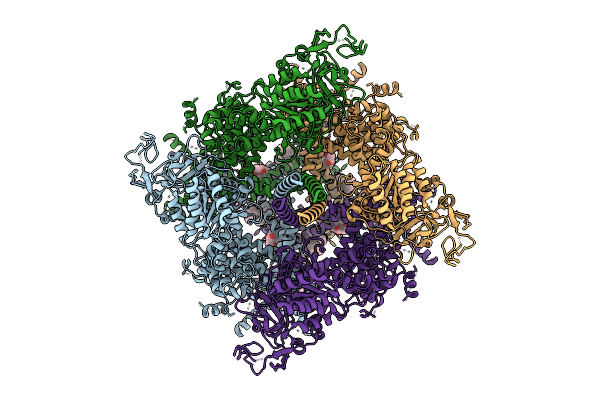
Cryo-Em Structure Of Rabbit Trpm3 In Apo Activated State At 37 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
 |
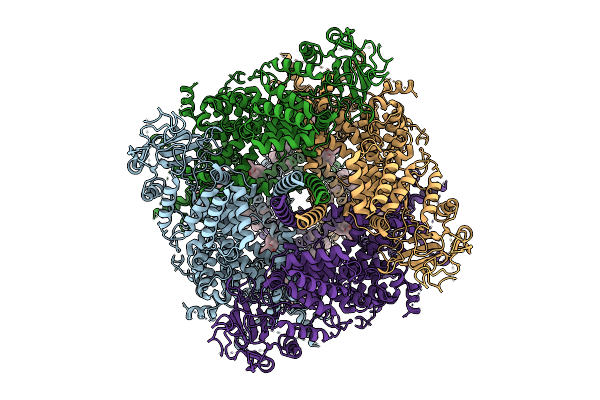
Cryo-Em Structure Of Rabbit Trpm3 In Complex With Primidone In Resting State At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01, A1AIA |
 |
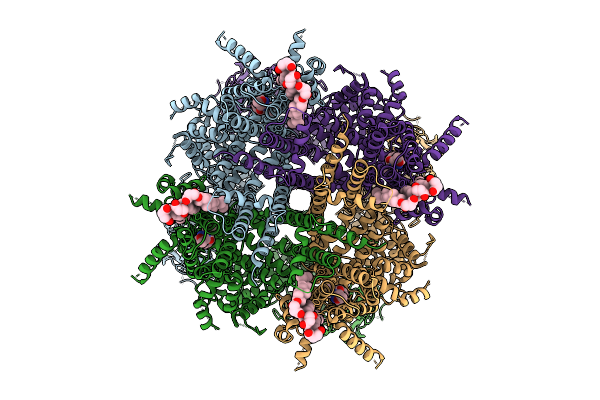
Cryo-Em Structure Of Rabbit Trpm3 In Complex With Primidone In Activated State At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01, A1AIA |
 |
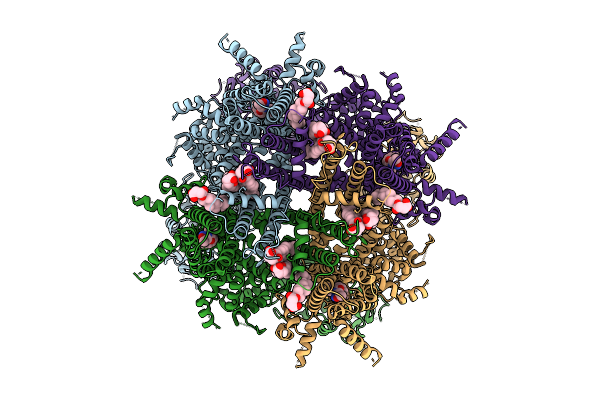
Cryo-Em Structure Of Rabbit Trpm3 In Complex With Cim0216 In Activated State At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01, A1AIB |
 |
Cryo-Em Structure Of Rabbit Trpm3 In Complex With Cim0216 At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01, A1AIB |
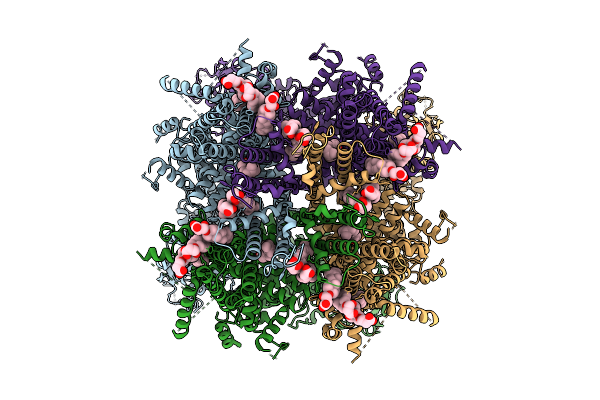 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 3 Resting And 1 Activated Subunits At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
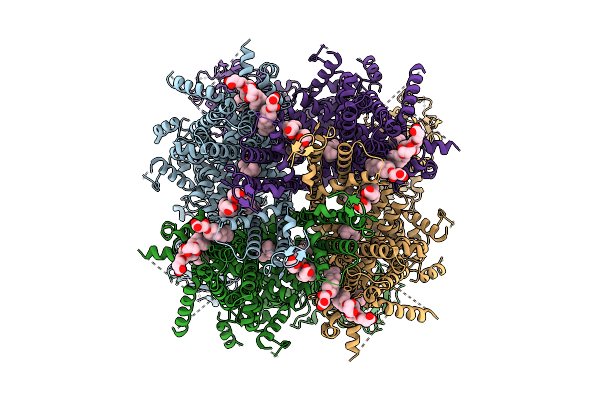 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 2 Resting And 2 Activated Subunits (Ortho Position) At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
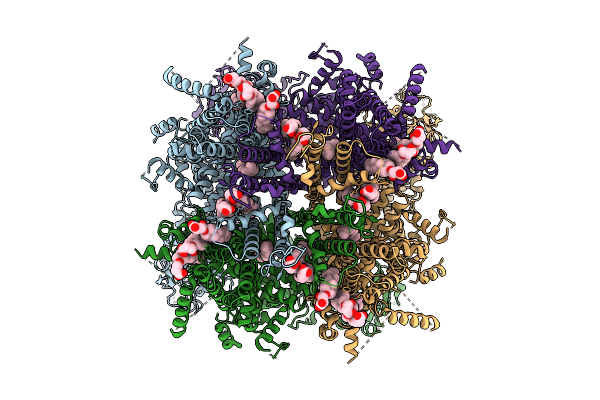 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 2 Resting And 2 Activated Subunits (Para Position) At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
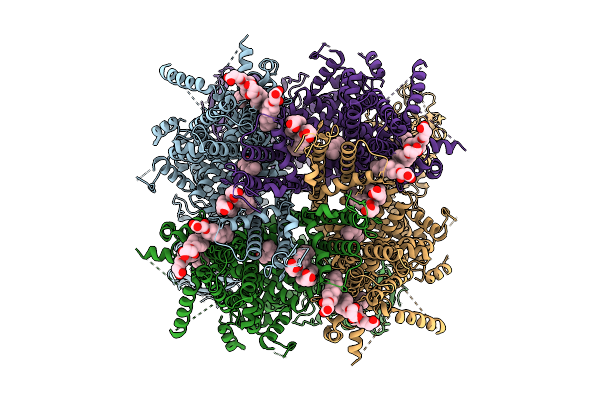 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 1 Resting And 3 Activated Subunits At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 3 Resting And 1 Activated Subunits At 37 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 2 Resting And 2 Activated Subunits (Ortho Position) At 37 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 2 Resting And 2 Activated Subunits (Para Position) At 37 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Apo Rabbit Trpm3 Having 1 Resting And 3 Activated Subunits At 37 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: Y01 |
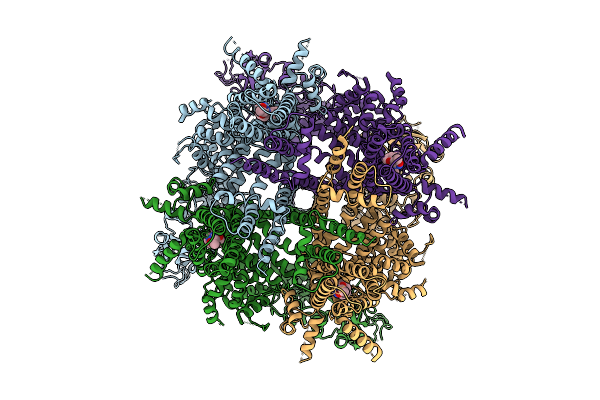 |
Cryo-Em Structure Of Cim0216-Bound Rabbit Trpm3 Having 3 Resting And 1 Activated Subunits At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: A1AIB |
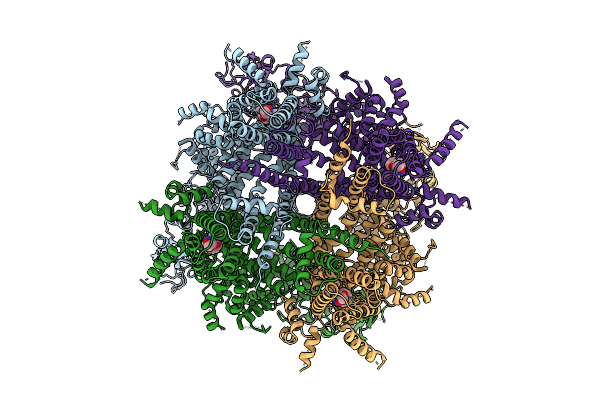 |
Cryo-Em Structure Of Cim0216-Bound Rabbit Trpm3 Having 2 Resting And 2 Activated Subunits (Ortho Position) At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: A1AIB |
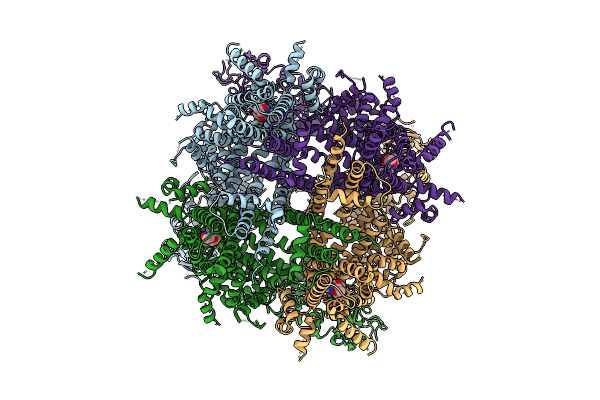 |
Cryo-Em Structure Of Cim0216-Bound Rabbit Trpm3 Having 2 Resting And 2 Activated Subunits (Para Position) At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: A1AIB |
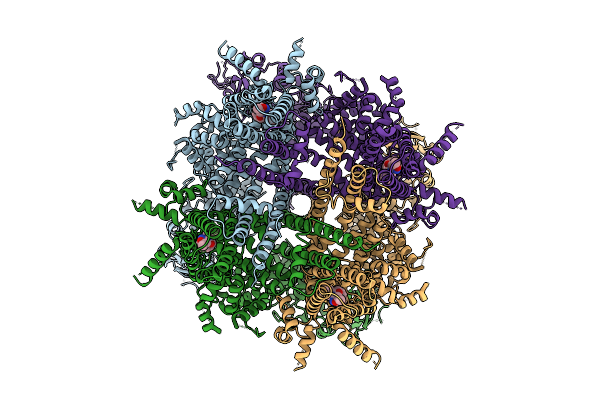 |
Cryo-Em Structure Of Cim0216-Bound Rabbit Trpm3 Having 1 Resting And 3 Activated Subunits At 18 Degrees Celsius
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: A1AIB |

