Search Count: 35
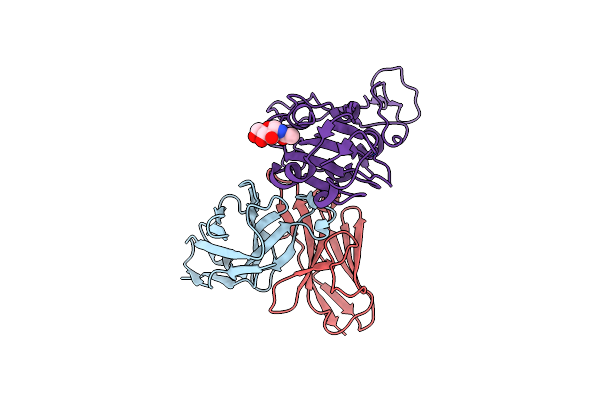 |
Sars-Cov-2 Spike Omicron (Ba.1) Ectodomain Trimer In Complex With Sc27 Fab, Local Refinement
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
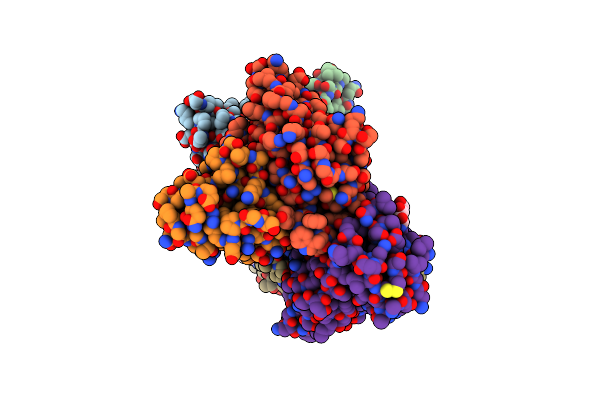 |
Sars-Cov-2 Spike Omicron (Ba.1) Rbd Ectodomain Dimer-Of-Trimers In Complex With Sc27 Fabs
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Cytaeis uchidae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-07-19 Classification: FLUORESCENT PROTEIN Ligands: CL, EDO |
 |
Organism: Homo sapiens, Zika virus zikv/h. sapiens/frenchpolynesia/10087pf/2013
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: VIRUS |
 |
Organism: Zika virus zikv/human/cambodia/fss13025/2010, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2023-01-04 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens, Zika virus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: VIRUS |
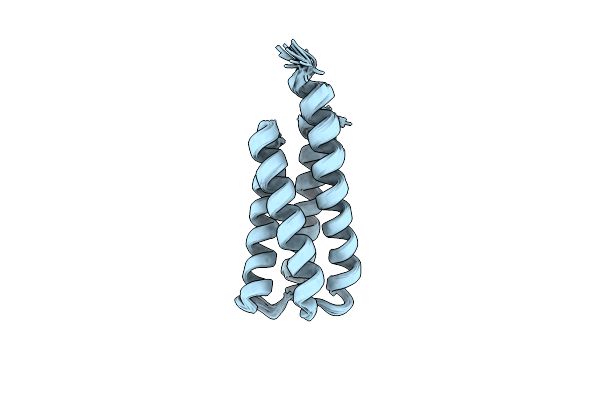 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2022-03-02 Classification: DE NOVO PROTEIN |
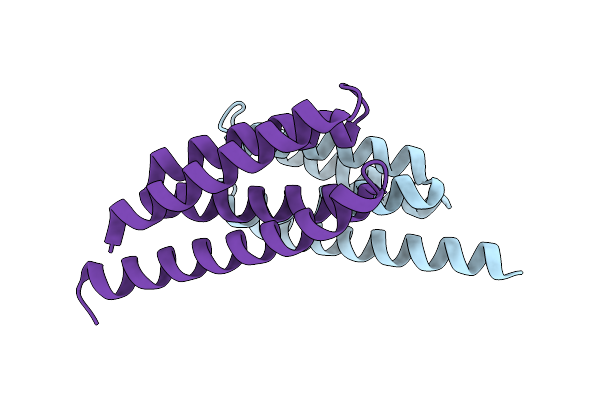 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2022-03-02 Classification: DE NOVO PROTEIN |
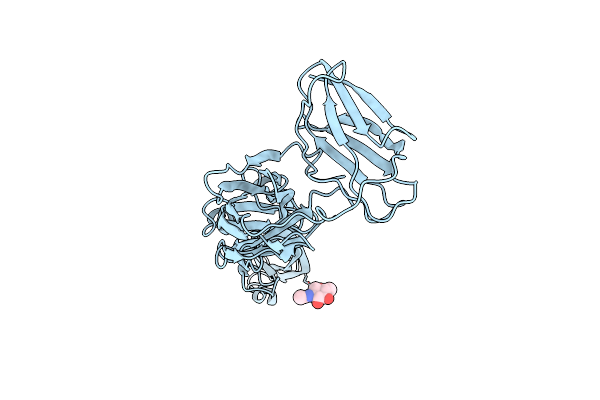 |
Crystal Structure Of An Engineered Thermostable Dengue Virus 2 Envelope Protein Dimer
Organism: Dengue virus 2
Method: X-RAY DIFFRACTION Resolution:3.42 Å Release Date: 2021-11-10 Classification: VIRAL PROTEIN Ligands: NAG |
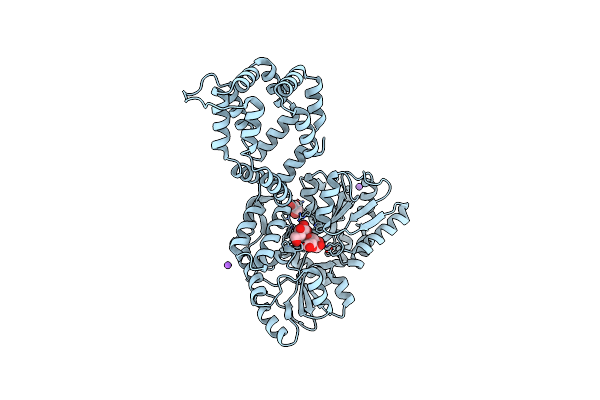 |
Organism: Escherichia coli, Aedes aegypti
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-03-24 Classification: LIPID BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-05-15 Classification: REPLICATION |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2019-05-15 Classification: REPLICATION |
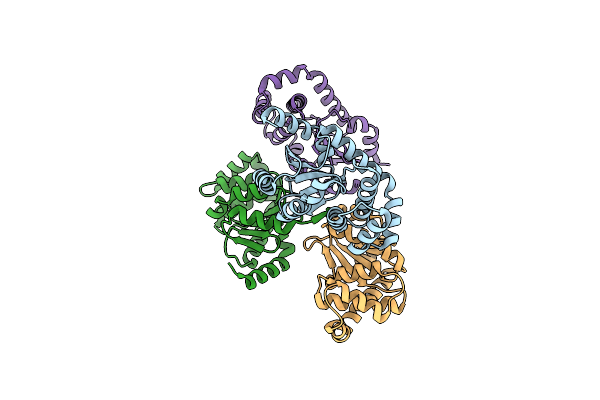 |
Crystal Structure Of The Thiol-Disulfide Exchange Protein Alpha-Dsba2 From Wolbachia Pipientis
Organism: Wolbachia endosymbiont of drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-04-17 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-03-06 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-03-06 Classification: OXIDOREDUCTASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2017-08-02 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2017-07-26 Classification: ISOMERASE |
 |
Organism: Proteus mirabilis atcc 29906
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-06-08 Classification: ISOMERASE |
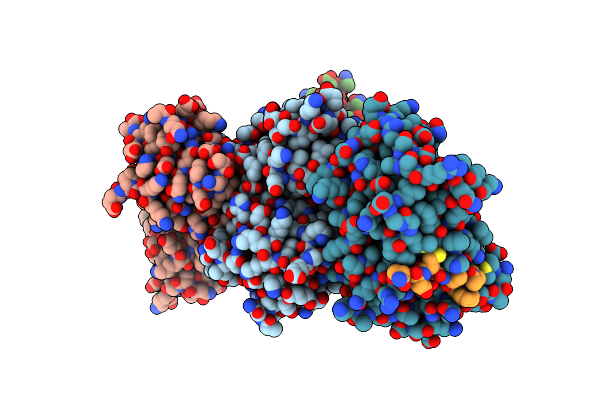 |
The Complex Structure Of E. Coli Dsba Bound To A Peptide At The Dsba/Dsbb Interface
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-01-14 Classification: OXIDOREDUCTASE/PEPTIDE INHIBITOR Ligands: ACE, NH2 |
 |
Organism: Acinetobacter baumannii aye, Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-06-18 Classification: TRANSLATION/OXIDOREDUCTASE Ligands: MG, GDP, GOL |

