Search Count: 42
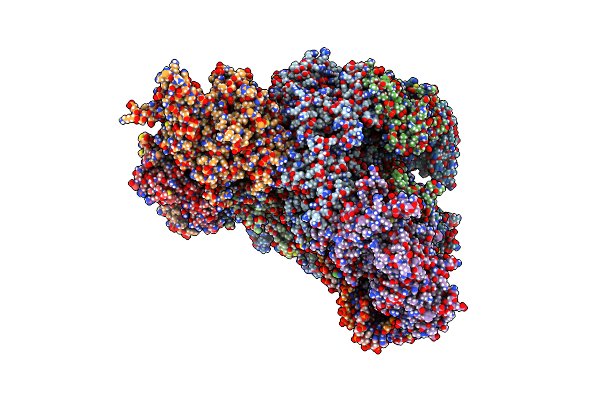 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 1
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: REPLICATION/DNA Ligands: ADP, MG, ZN, ATP |
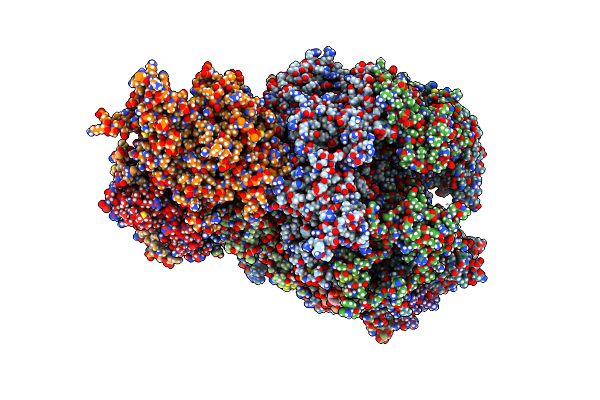 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 3
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: REPLICATION/DNA Ligands: ADP, MG, ZN, ATP |
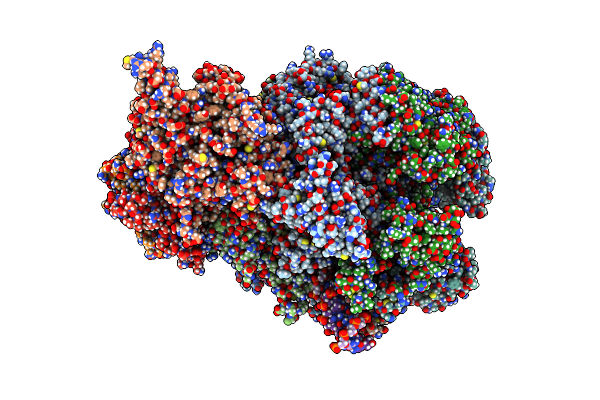 |
Cryo-Em Structure Of Human Cmg Helicase Stalled At G4-Containing Dna Template
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: REPLICATION/DNA Ligands: ADP, MG, ATP, ZN, K |
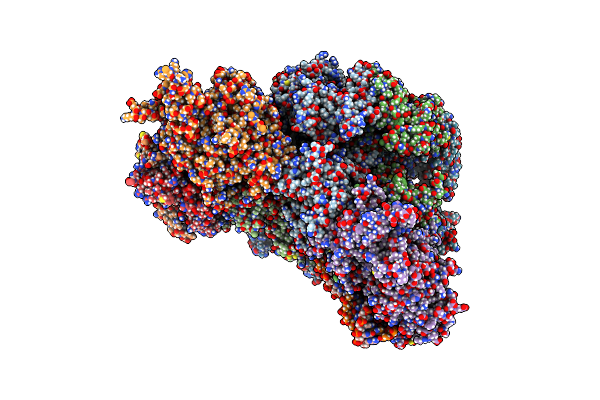 |
Cryo-Em Structure Of Yeast Cmg Helicase Stalled At G4-Containing Dna Template, State 2
Organism: Saccharomyces cerevisiae w303, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-19 Classification: REPLICATION/DNA Ligands: ATP, MG, ZN, ADP |
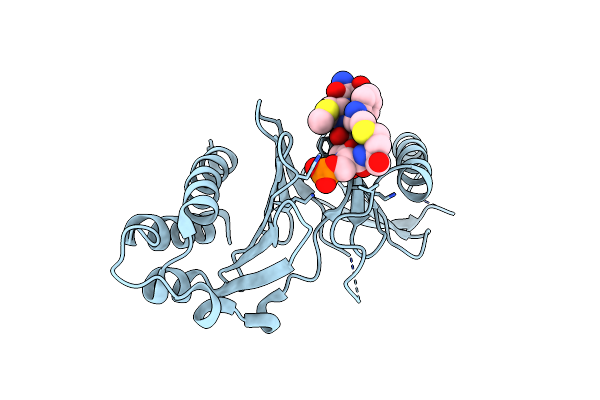 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
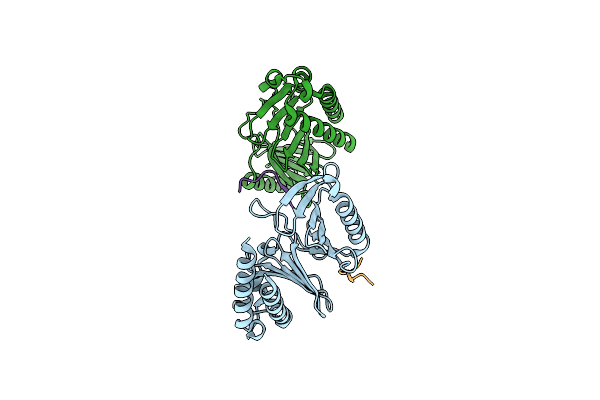 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2024-08-21 Classification: CELL CYCLE |
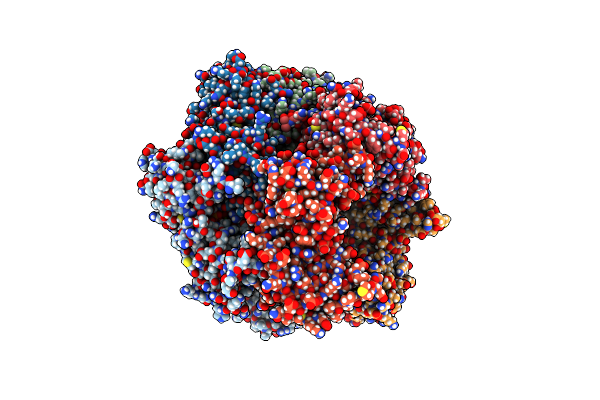 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
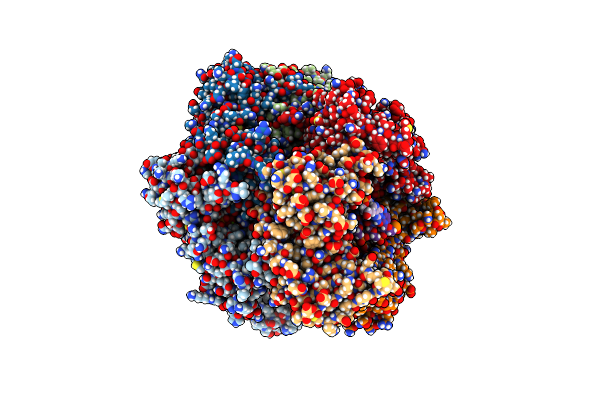 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-24 Classification: REPLICATION/DNA Ligands: AGS, MG, ADP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: REPLICATION/DNA Ligands: MG, AGS, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: REPLICATION/DNA Ligands: MG, AGS, GDP |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-08-17 Classification: REPLICATION/DNA Ligands: AGS, MG, GDP |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand (2R)-2-Amino-N-[3-(Difluorom Ethoxy)-4-(1,3-Oxazol-5-Yl)Phenyl]-4-Methylpentanamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-02-23 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: YFV, SO4, 5OI |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand 2-(1-Amino-3-Methylbutyl)-6- (Pyridin-4-Yl)Quinoline-4-Carbonitrile
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2022-02-23 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5P6, SO4 |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand (2R)-2-Amino-N-[3-(Difluorom Ethoxy)-4-(1,3-Oxazol-5-Yl)Phenyl]-4-Methylpentanamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2022-02-23 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5QI, PG6, SO4, YFV |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Isoform 1 Complexed With Ligand N-[3-Methoxy-4-(1,3-Oxazol-5-Yl)Phenyl]-3-(Propan-2-Yl)Piperidine-2-Carboxamide
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2021-07-28 Classification: TRANSFERASE/TRANSFERASE inhibitor Ligands: YFS, SO4, YFV |

