Search Count: 15
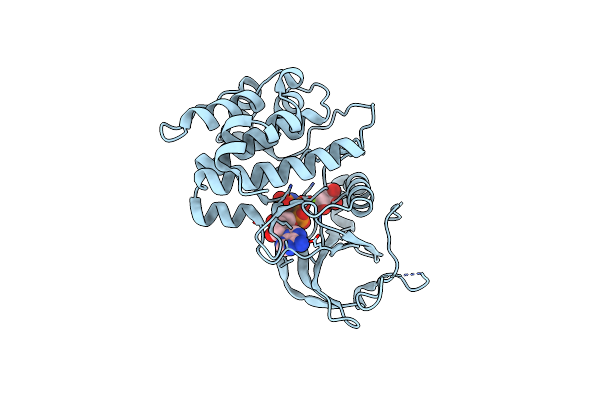 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 In Complex With Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: ANP, GOL, CL, MG |
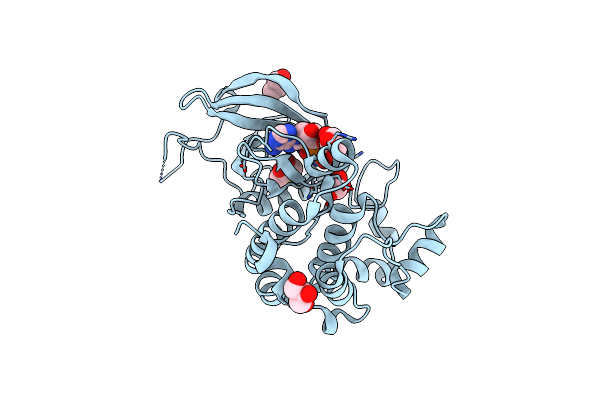 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 Modified With Cacodylate And In Complex With Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: CL, DTD, ANP, GOL |
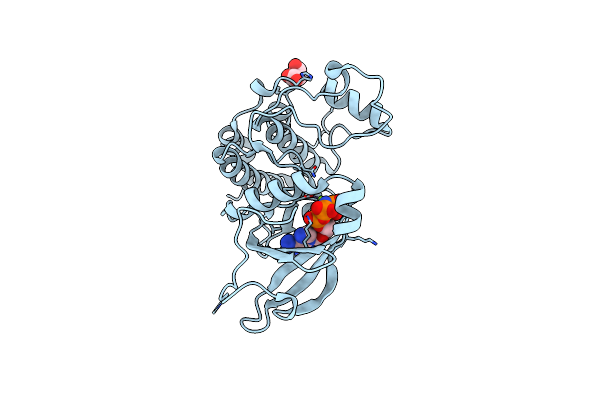 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 C247V + D256N + C319V Triple Mutant Disulfide Homodimer In Complex With Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: ANP, MG, MLA |
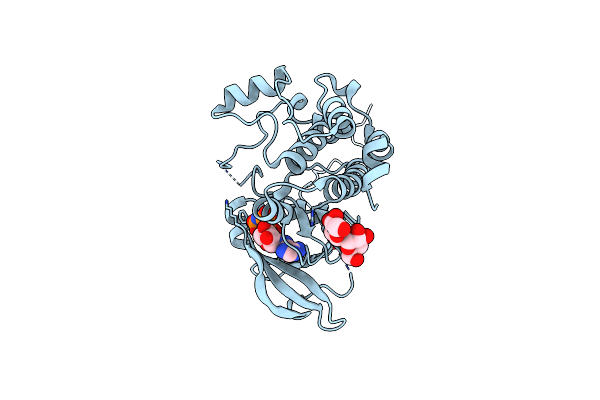 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 C247V + C319V Double Mutant Dephosphorylated, And In Complex With Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: ANP |
 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 With C290 Disulfide Bonded To Compound 7-80, And In Complex With Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: MG, ANP, MLA, R7D, GOL |
 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 With C290 Disulfide Bonded To Compound 8-34, And In Complex With Amp-Pnp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: MG, AMP, R74, MLA |
 |
Tpx2 Residues 7-20 Fused To Aurora A Residues 116-389 Dephosphorylated, And Coalated On C290
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-08-05 Classification: TRANSFERASE Ligands: COA |
 |
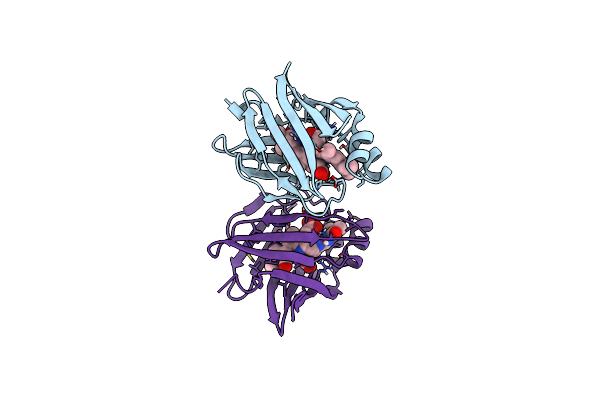
Organism: Anguilla japonica
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2013-06-19 Classification: FLUORESCENT PROTEIN Ligands: BLR, PEG |
 |
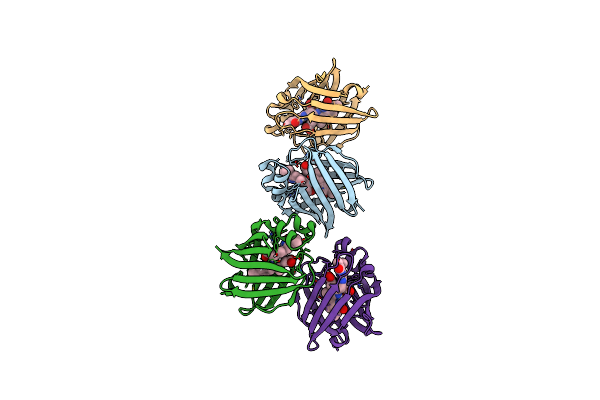
Organism: Anguilla japonica
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-06-19 Classification: FLUORESCENT PROTEIN Ligands: BLR |
 |
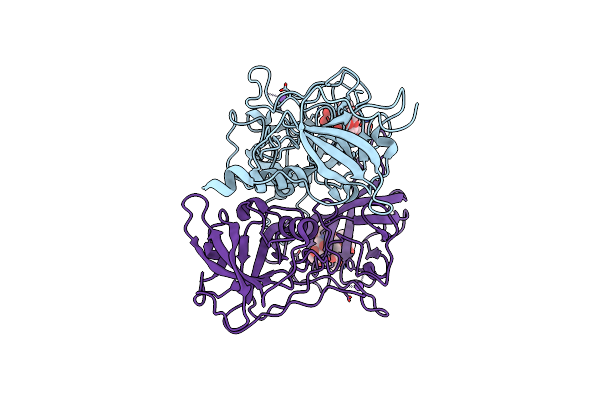
Organism: Anguilla japonica
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-06-19 Classification: FLUORESCENT PROTEIN Ligands: BLR |
 |
Crystal Structure Of P Domain From Norovirus Funabashi258 Stain In The Complex With A-Antigen
Organism: Norwalk-like virus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-01-25 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of P Domain From Norovirus Funabashi258 Stain In The Complex With H-Antigen
Organism: Norwalk-like virus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-01-25 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of P Domain From Norovirus Funabashi258 Stain In The Complex With Lewis-A
Organism: Norwalk-like virus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-01-25 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of P Domain From Norovirus Funabashi258 Stain In The Complex With Lewis-B
Organism: Norwalk-like virus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-01-25 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of P Domain Q389N Mutant From Norovirus Funabashi258 Stain In The Complex With Lewis-B
Organism: Norwalk-like virus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-01-25 Classification: VIRAL PROTEIN |

