Search Count: 5
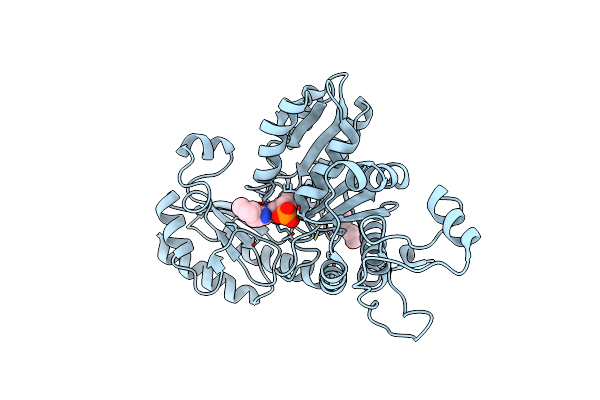 |
Crystal Structure Of Citrobacter Freundii Methionine Gamma-Lyase With C115H Replacement In The Complex With L-Norleucine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-06-28 Classification: LYASE Ligands: PLP, NLE, PGE, PY6 |
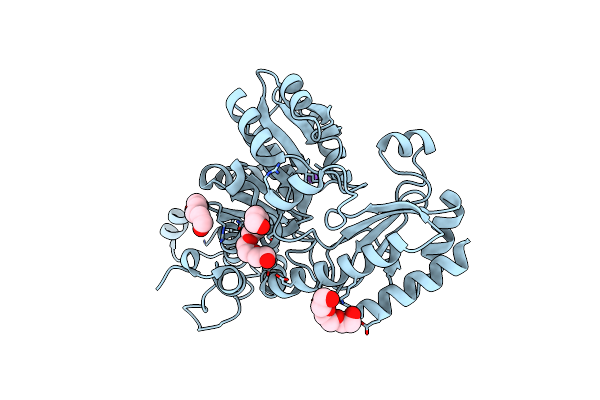 |
Crystal Structure Of L-Methionine Gamma-Lyase From Citrobacter Freundii Modified By Allicine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-11-12 Classification: LYASE Ligands: 1PE, PGE, NA |
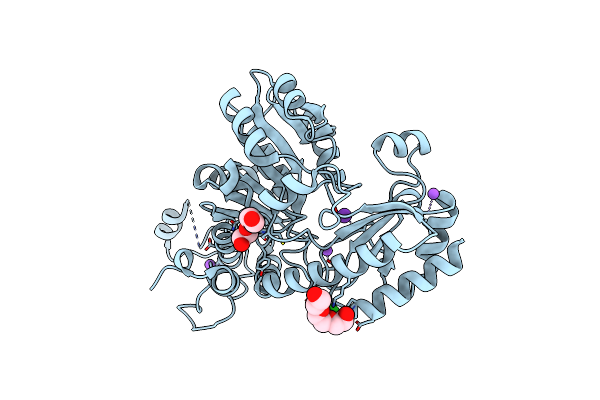 |
Crystal Structure Of C115A Mutant L-Methionine Gamma-Lyase From Citrobacter Freundii Modified By Allicine
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2014-11-12 Classification: LYASE Ligands: PGE, NA, CL, K |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-07-25 Classification: LYASE Ligands: K |
 |
Organism: Citrobacter freundii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-07-25 Classification: LYASE Ligands: K, PO4 |

