Search Count: 7
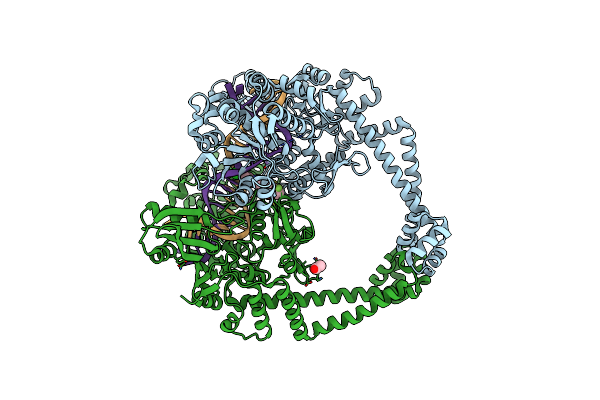 |
Crystal Structure Of S. Aureus Gyrase In Complex With 4-[[1-[(1-Chloro-6,7-Dihydro-5H-Cyclopenta[C]Pyridin-6-Yl)Methyl]Azetidin-3-Yl]Methylamino]-6-Fluorochromen-2-One
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-06-28 Classification: ISOMERASE Ligands: MN, CL, ACT, 61I |
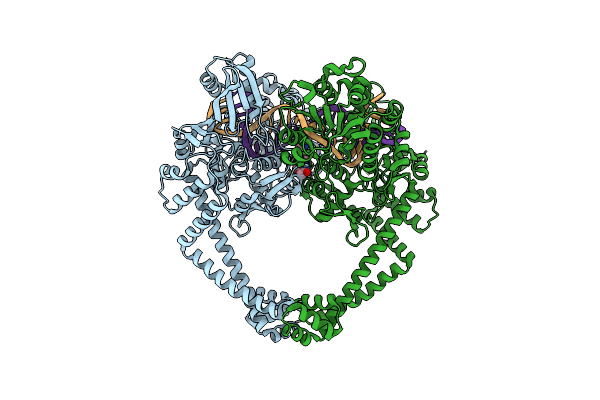 |
Crystal Structure Of S. Aureus Gyrase In Complex With 6-[5-[2-[(4-Chloro-2,3-Dihydro-1H-Inden-2-Yl)Methylamino]Ethyl]-2-Oxo-1,3-Oxazolidin-3-Yl]-4H-Pyrido[3,2-B][1,4]Oxazin-3-One
Organism: Staphylococcus aureus subsp. aureus n315, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2023-06-28 Classification: ISOMERASE Ligands: MN, CL, 6I0 |
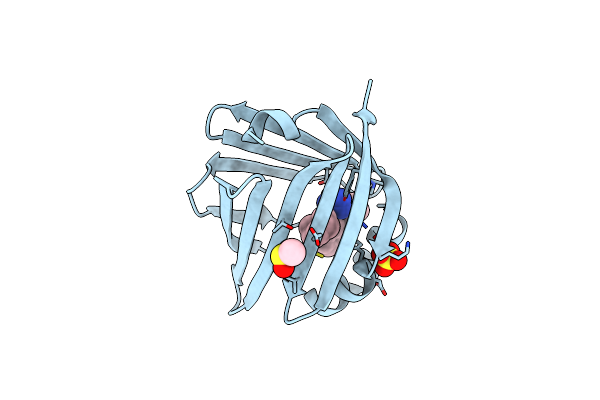 |
Fabp5 In Complex With 6-Chloro-4-Phenyl-2-Piperidin-1-Yl-3-(1H-Tetrazol-5-Yl)-Quinoline
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-01-25 Classification: LIPID BINDING PROTEIN Ligands: DMS, SO4, 65X |
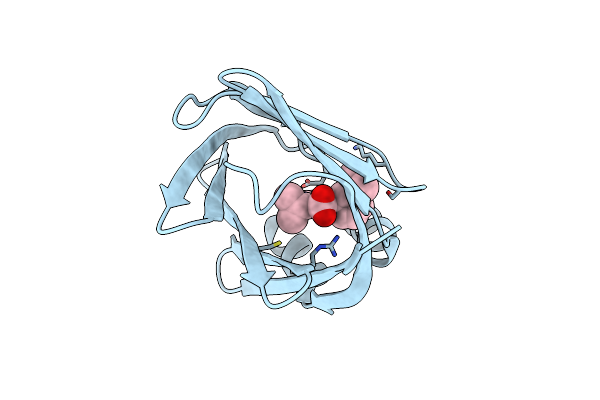 |
Fabp4 In Complex With 6-Chloro-2-Isopropyl-4-(3-Isopropyl-Phenyl)-Quinoline-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: 65Y |
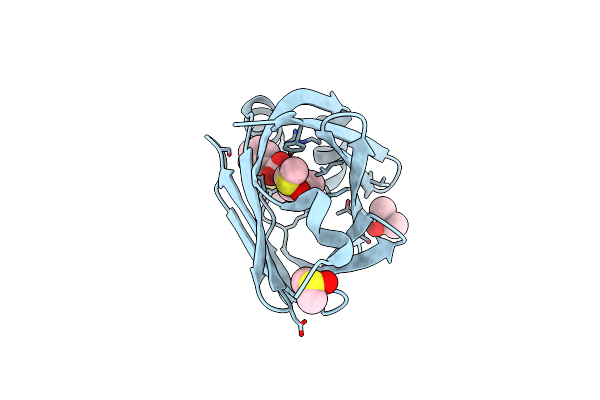 |
Fabp4_3 In Complex With 6,8-Dichloro-4-Phenyl-2-Piperidin-1-Yl-Quinoline-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2016-12-14 Classification: TRANSPORT PROTEIN Ligands: 65Z, DMS |
 |
Human Fabp3 In Complex With 6-Chloro-2-Methyl-4-Phenyl-Quinoline-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-12-14 Classification: LIPID BINDING PROTEIN Ligands: SO4, 5M8, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-10-25 Classification: HYDROLASE Ligands: GOL, CA, 413, SO4 |

