Search Count: 11
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Homocysteine Bound.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Methionine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAM, EDO, GOL |
 |
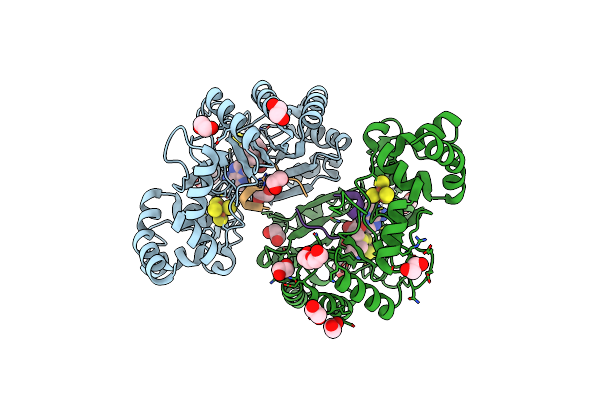
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG, GOL |
 |
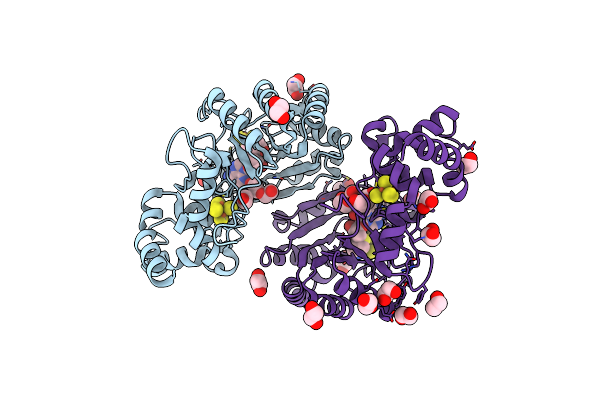
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 6 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
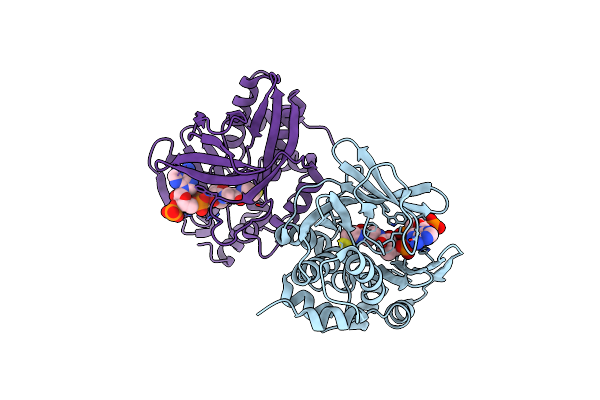
Crystal Structure Of Radical Sam Epimerase Epee D210A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Persulfurated Cysteine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
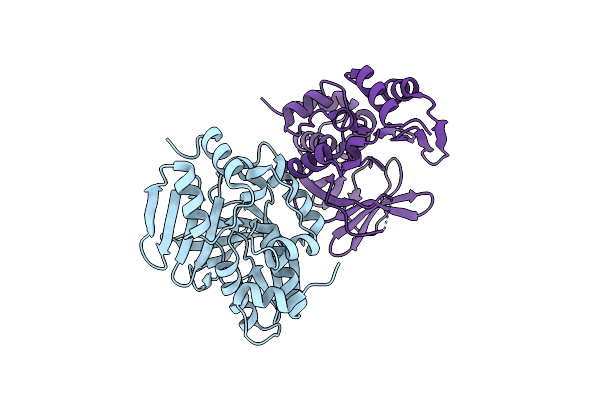
Crystal Structure Of Mesorhizobium Loti Arylamine N-Acetyltransferase 1 In Complex With Coa
Organism: Mesorhizobium loti
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2015-01-21 Classification: TRANSFERASE Ligands: COA |
 |
Crystal Structure Of Mesorhizobium Loti Arylamine N-Acetyltransferase F42W Mutant
Organism: Mesorhizobium loti
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-01-21 Classification: TRANSFERASE |
 |
Structure Of The Arylamine N-Acetyltransferase From Mycobacterium Abscessus
Organism: Mycobacterium abscessus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-10-23 Classification: TRANSFERASE |
 |
Crystal Structure Of The (Baccr)Nat3 Arylamine N-Acetyltransferase From Bacillus Cereus Reveals A Unique Cys-His-Glu Catalytic Triad
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2013-06-26 Classification: TRANSFERASE Ligands: KH2 |
 |
Crystal Structure Analysis Of Arylamine N-Acetyltransferase C From Bacillus Anthracis
Organism: Bacillus anthracis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2011-01-26 Classification: TRANSFERASE Ligands: COA, FMT |

