Search Count: 20
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-01-11 Classification: Transferase/Transferase Inhibitor Ligands: 7CU |
 |
Crystal Structure Of Evaa, A 2,3-Dehydratase In Complex With Dtdp-Fucose And Dtdp-Rhamnose
Organism: Amycolatopsis orientalis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-05-22 Classification: BIOSYNTHETIC PROTEIN Ligands: TRH, 1JB |
 |
Crystal Structure Of Evaa, A 2,3-Dehydratase In Complex With Dtdp-Benzene And Dtdp-Rhamnose
Organism: Amycolatopsis orientalis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2013-05-22 Classification: BIOSYNTHETIC PROTEIN Ligands: TLO, TRH, EDO |
 |
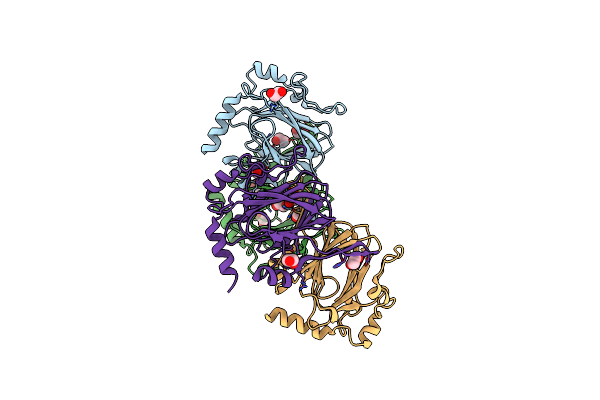
Crystal Structure Of Chmj, A 3'-Monoepimerase From Streptomyces Bikiniensis In Complex With Dtdp-Quinovose
Organism: Streptomyces bikiniensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-11-21 Classification: UNKNOWN FUNCTION Ligands: EDO, 18T |
 |
Crystal Structure Of Chmj, A 3'-Monoepimerase Apoenzyme From Streptomyces Bikiniensis
Organism: Streptomyces bikiniensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-11-21 Classification: UNKNOWN FUNCTION Ligands: EDO, CL |
 |
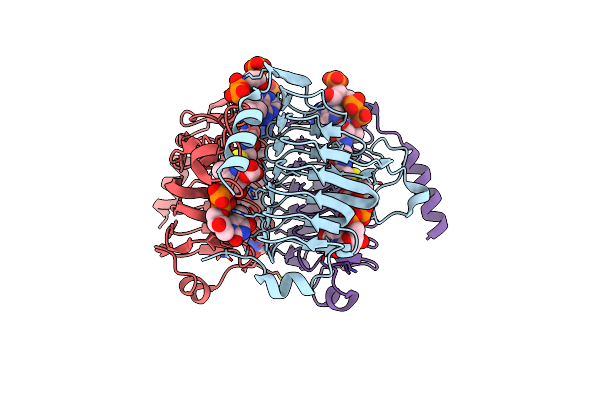
Crystal Structure Of H60N/Y130F Double Mutant Of Chmj, A 3'-Monoepimerase From Streptomyces Bikiniensis In Complex With Dtdp
Organism: Streptomyces bikiniensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-11-21 Classification: UNKNOWN FUNCTION Ligands: EDO, TYD, THM, TDR |
 |
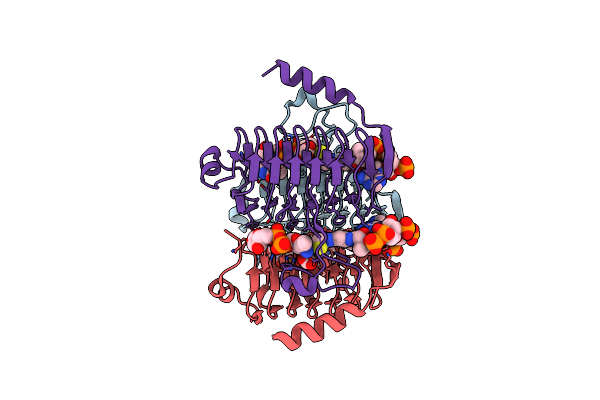
Crystal Structure Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp-4-Amino-4,6-Dideoxyglucose And Coenzyme A
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, 0FX, BCT, CL |
 |
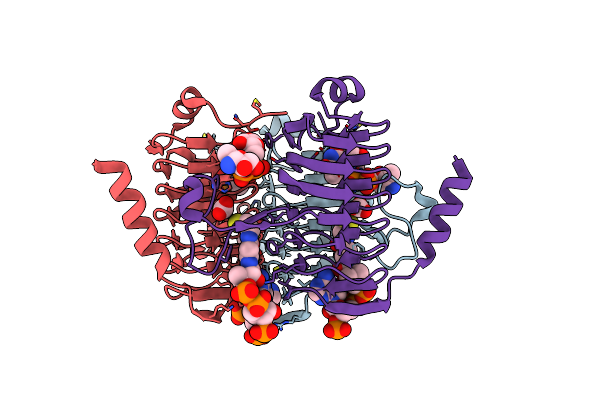
Crystal Structure Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp And 3-Hydroxybutyryl-Coa
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: CL, TYD, 3HC |
 |
Crystal Structure Of The S84A Mutant Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp-4-Amino-4,6-Dideoxyglucose And Coenzyme A
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, 0FX, CL, BCT |
 |
Crystal Structure Of The S84C Mutant Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp-4-Amino-4,6-Dideoxyglucose And Coenzyme A
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, CL, 0FX, BCT |
 |
Crystal Structure Of The S84T Mutant Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp And Coenzyme A
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, TYD, BCT, CL |
 |
Crystal Structure Of The D94A Mutant Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp And Coenzyme A
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, TYD, CL |
 |
Crystal Structure Of The D94N Mutant Of Antd, An N-Acyltransferase From Bacillus Cereus In Complex With Dtdp And Coenzyme A
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-01-25 Classification: TRANSFERASE Ligands: COA, TYD, CL |
 |
Crystal Structure Of Kijd10, A 3-Ketoreductase From Actinomadura Kijaniata Incomplex With Nadp
Organism: Actinomadura kijaniata
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-08 Classification: SUGAR BINDING PROTEIN Ligands: EDO, NAP, CL, NA, PO4 |
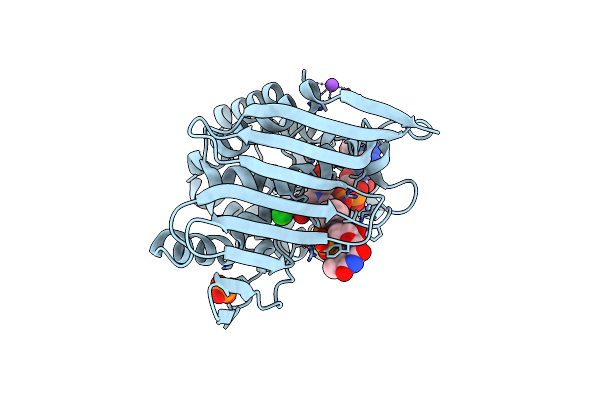 |
Crystal Structure Of Kijd10, A 3-Ketoreductase From Actinomadura Kijaniata Incomplex With Nadp And Tdp-Benzene
Organism: Actinomadura kijaniata
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2011-06-08 Classification: SUGAR BINDING PROTEIN Ligands: PO4, TLO, NAP, NA, EDO, CL |
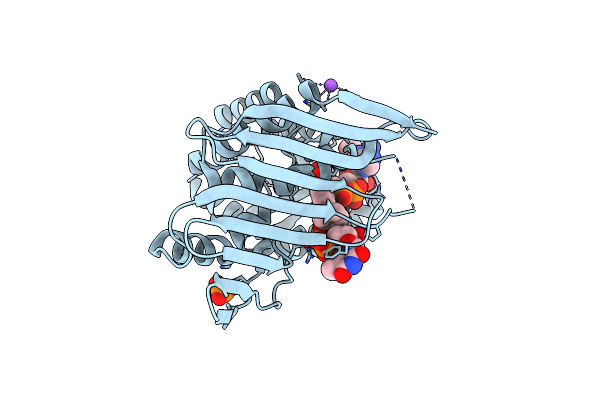 |
Crystal Structure Of Kijd10, A 3-Ketoreductase From Actinomadura Kijaniata In Complex With Tdp-Benzene And Nadp; Open Conformation
Organism: Actinomadura kijaniata
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-06-08 Classification: SUGAR BINDING PROTEIN Ligands: PO4, EDO, TLO, NAP, CL, NA |
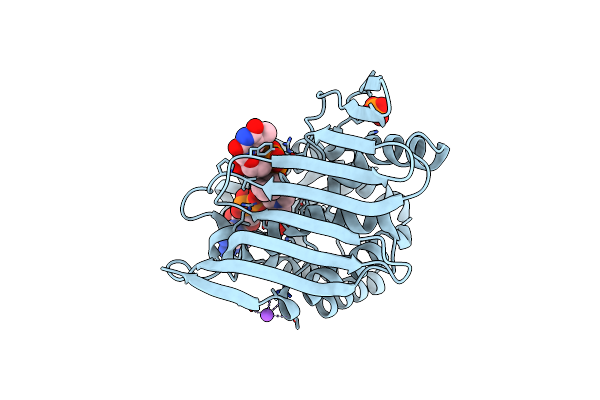 |
Crystal Structure Of The Y186F Mutant Of Kijd10, A 3-Ketoreductase From Actinomadura Kijaniata In Complex With Tdp-Benzene And Nadp
Organism: Actinomadura kijaniata
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-06-08 Classification: SUGAR BINDING PROTEIN Ligands: NAP, CL, NA, PO4, TLO |
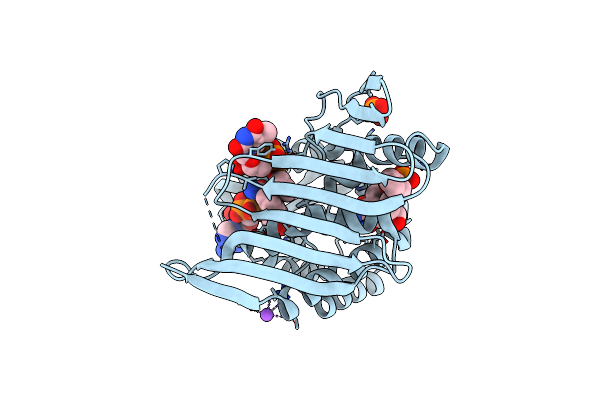 |
Crystal Structure Of The K102A Mutant Of Kijd10, A 3-Ketoreductase From Actinomadura Kijaniata In Complex With Tdp-Benzene And Nadp
Organism: Actinomadura kijaniata
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2011-06-08 Classification: SUGAR BINDING PROTEIN Ligands: NAP, CL, NA, PO4, TLO |
 |
Crystal Structure Of The K102E Mutant Of Kijd10, A 3-Ketoreductase From Actinomadura Kijaniata In Complex With Tdp-Benzene And Nadp
Organism: Actinomadura kijaniata
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2011-06-08 Classification: SUGAR BINDING PROTEIN Ligands: NAP, CL, NA, PO4, TLO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-06-16 Classification: TRANSFERASE Ligands: AKG |

