Search Count: 91
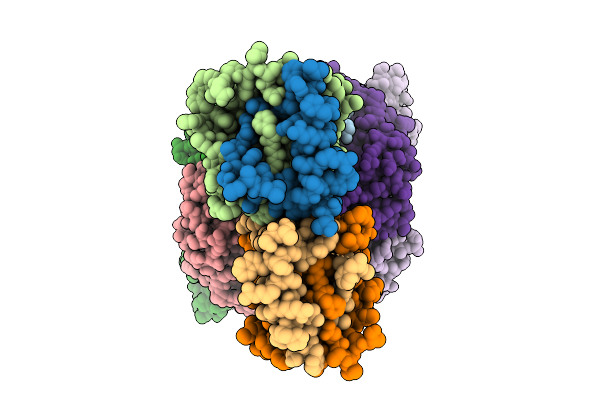 |
Thermus Thermophilus Mrec-Mred Complex With An Internal Mred Bril Fusion And An Anti-Bril Fab
Organism: Escherichia coli, Thermus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
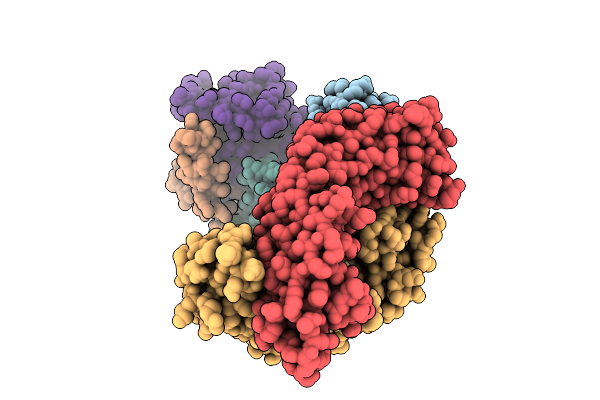 |
Thermus Thermophilus Mrec-Mred Complex With A C-Terminal Mred Bril Fusion And An Anti-Bril Fab
Organism: Escherichia coli, Thermus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
 |
Organism: Rhodobacteraceae bacterium qy30, Dna molecule
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN/DNA Ligands: MN |
 |
Organism: Rhodobacteraceae bacterium qy30
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN/DNA Ligands: PO4 |
 |
Organism: Rhodobacteraceae bacterium qy30
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN Ligands: A1A3G, F2A, MN, A1A3H, D5M |
 |
Organism: Rhodobacteraceae bacterium qy30, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN |
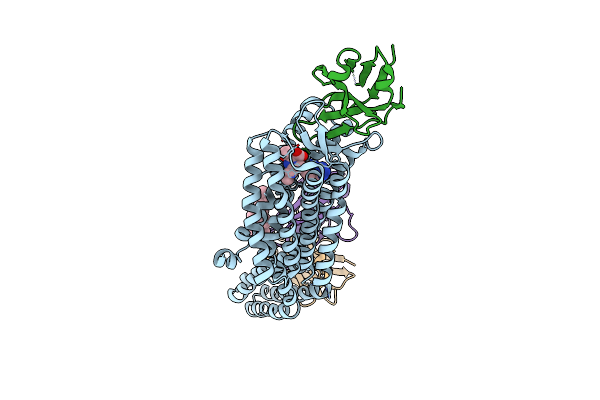 |
Structure Of Nanobody At209 In Complex With The Olmesartan-Bound Angiotensin Ii Type I Receptor (At1R)
Organism: Camelidae, Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: OLM, CLR |
 |
Structure Of Nanobody At206 In Complex With The Losartan-Bound Angiotensin Ii Type I Receptor (At1R)
Organism: Camelidae, Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN Ligands: LSN, Y01, CLR |
 |
Structure Of Nanobody At206 In Complex With The Angiotensin Ii Type I Receptor (At1R)
Organism: Camelidae, Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: MEMBRANE PROTEIN |
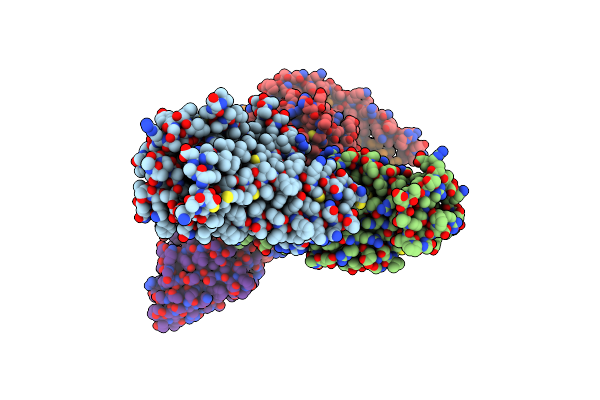 |
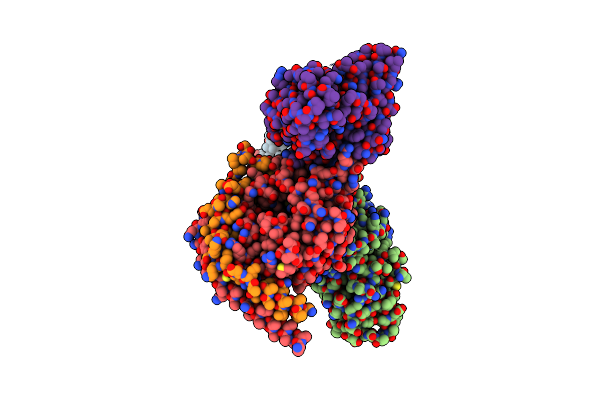
5-Ht2Ar Bound To Lisuride In Complex With A Mini-Gq Protein And An Active-State Stabilizing Single-Chain Variable Fragment (Scfv16) Obtained By Cryo-Electron Microscopy (Cryoem)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANE PROTEIN Ligands: H8G |
 |
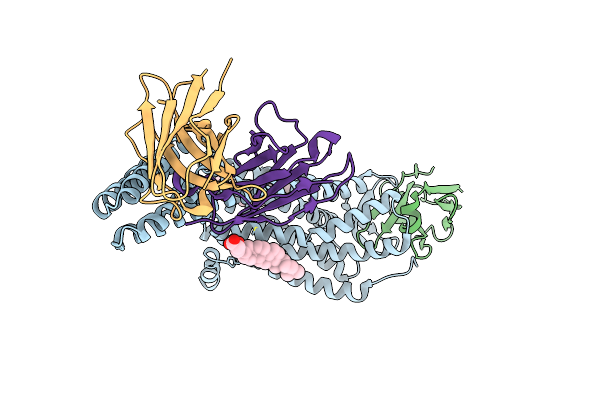
5Ht2Ar-Minigq Heterotrimer In Complex With A Novel Agonist Obtained From Large Scale Docking
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: MEMBRANE PROTEIN Ligands: YEQ |
 |
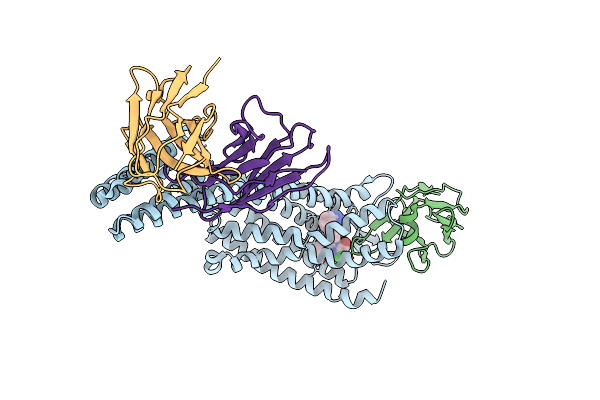
Structure Of At118-H Nanobody Antagonist In Complex With The Angiotensin Ii Type I Receptor
Organism: Camelidae, Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
 |
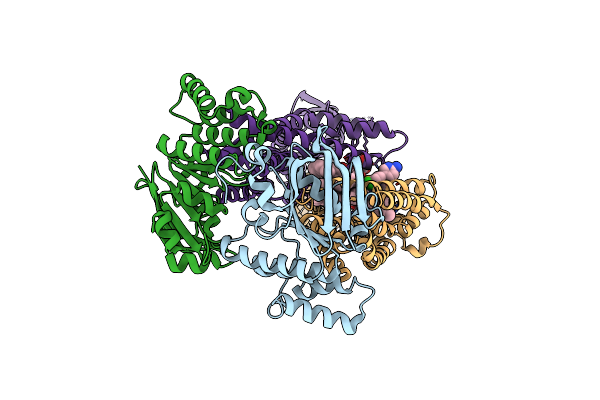
Structure Of At118-L Nanobody Antagonist In Complex With The Angiotensin Ii Type I Receptor And Losartan
Organism: Homo sapiens, Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: LSN |
 |
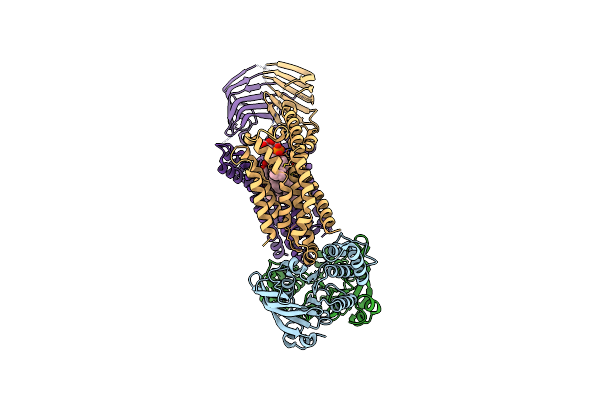
Acinetobacter Baylyi Lptb2Fg Bound To Lipopolysaccharide And A Macrocyclic Peptide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: Y75, JSG, LMT |
 |
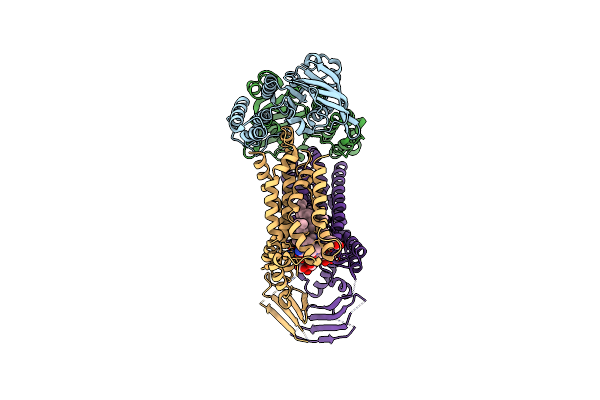
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: JSG |
 |
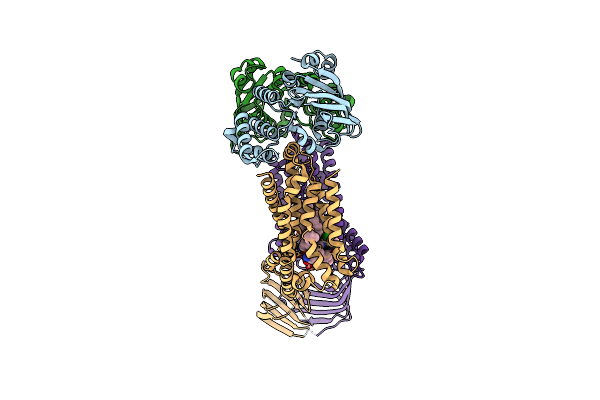
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: JSG, VB6, LMT |
 |
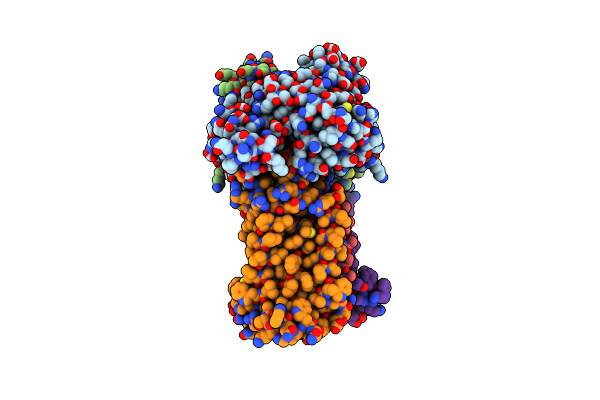
Acinetobacter Baylyi Lptb2Fg Bound To Lipopolysaccharide And A Macrocyclic Peptide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: JSG, MG3 |
 |
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT |
 |
Acinetobacter Baylyi Lptb2Fg Bound To Acinetobacter Baylyi Lipopolysaccharide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: LMT, WJR |
 |
Acinetobacter Baylyi Lptb2Fg Bound To Acinetobacter Baylyi Lipopolysaccharide And A Macrocyclic Peptide
Organism: Acinetobacter baylyi adp1
Method: ELECTRON MICROSCOPY Release Date: 2024-01-03 Classification: LIPID TRANSPORT Ligands: WJW, Y75 |

