Search Count: 7
 |
Crystal Structure Of Acyl-Coa Dehydrogenase Fade2 Mutant (Pa0508 M130G E296A Q303A) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2025-02-19 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PGE |
 |
Crystal Structure Of The Acyl-Coa Dehydrogenase Fade1(Pa0506) E441A From Pseudomonas Aeruginosa Complexed With C16Coa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: PKZ, EDO, PGE, NO3, TRS |
 |
Crystal Structure Of The Apo Acyl-Coa Dehydrogenase Fade2 (Pa0508) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, 1PE, GOL |
 |
Crystal Structure Of The Acyl-Coa Dehydrogenase Pa0506 (Fade1) From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CO8 |
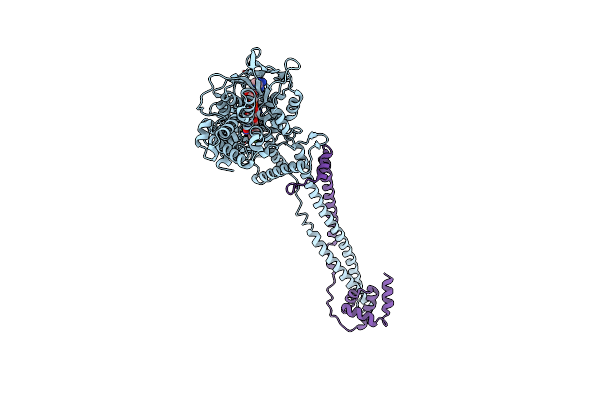 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2024-07-10 Classification: OXIDOREDUCTASE Ligands: A1IG2 |
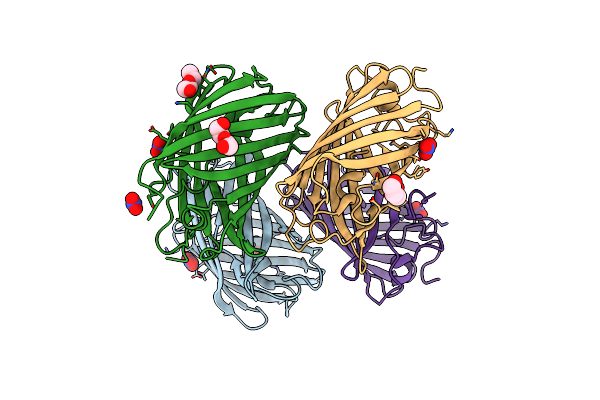 |
Crystal Structure Of A Reconstructed Kaede-Type Red Fluorescent Protein, Lea A69T
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-11-29 Classification: FLUORESCENT PROTEIN Ligands: NO3, PEG |
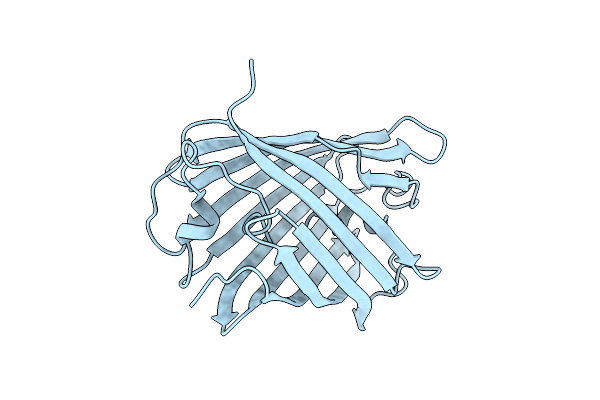 |
Crystal Structure Of A Reconstructed Kaede-Type Red Fluorescent Protein, Lea H62X, Containing 3-Methylhistidine At Position 62
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-11-29 Classification: LUMINESCENT PROTEIN |

