Search Count: 68
 |
Crystal Structure Of Mycobacterium Avium Dihydrofolate Reductase In Complex With Nadph And Trimethoprim
Organism: Mycobacterium avium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: NDP, TOP, CL |
 |
E. Coli Dihydrofolate Reductase Complexed With 5-(3-(7-(4-(Aminomethyl)Phenyl)Benzo[D][1,3]Dioxol-5-Yl)But-1-Yn-1-Yl)-6-Ethylpyrimidine-2,4-Diamine (Ucp1223)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2022-07-27 Classification: OXIDOREDUCTASE Ligands: SO4, 560 |
 |
Dfra1 Complexed With Nadph And 4'-Chloro-3'-(4-(2,4-Diamino-6-Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (Ucp1228)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2022-07-27 Classification: OXIDOREDUCTASE Ligands: 4SI, NAP, CA |
 |
Crystal Structure Of Escherichia Coli Dihydrofolate Reductase In Complex With Trimethoprim
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-29 Classification: OXIDOREDUCTASE Ligands: TOP, SO4 |
 |
Crystal Structure Of Dfra5 Dihydrofolate Reductase In Complex With Trimethoprim And Nadph
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2022-06-29 Classification: OXIDOREDUCTASE Ligands: TOP, NDP, SO4 |
 |
Crystal Structure Of Dfra1 Dihydrofolate Reductase In Complex With Trimethoprim
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: TOP |
 |
Crystal Structure Of Escherichia Coli Dihydrofolate Reductase In Complex With Trimethoprim And Nadph
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: ARG, NAP, TOP, SO4 |
 |
Dfra1 Complexed With Nadph And 5-(3-(7-(4-(Aminomethyl)Phenyl)Benzo[D][1,3]Dioxol-5-Yl)But-1-Yn-1-Yl)-6-Ethylpyrimidine-2,4-Diamine (Ucp1223)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2022-05-25 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAP, CA, 8MU, 560 |
 |
Dfra5 Complexed With Nadph And 5-(3-(7-(4-(Aminomethyl)Phenyl)Benzo[D][1,3]Dioxol-5-Yl)But-1-Yn-1-Yl)-6-Ethylpyrimidine-2,4-Diamine (Ucp1223)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-05-25 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAP, SO4, 560 |
 |
Dfra5 Complexed With Nadph And 4'-Chloro-3'-(4-(2,4-Diamino-6-Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (Ucp1228)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2022-05-25 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAP, GOL, 4SI, SO4 |
 |
E. Coli Dihydrofolate Reductase Complexed With 4'-Chloro-3'-(4-(2,4-Diamino-6-Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl)-[1,1'-Biphenyl]-4-Carboxamide (Ucp1228)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: ZM4, SO4 |
 |
Crystal Structure Of Escherichia Coli Enolase Complexed With A Natural Inhibitor Sf2312.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2019-11-27 Classification: LYASE/LYASE inhibitor Ligands: 4NG, MG, GOL, SO4 |
 |
Structure Of E.Coli Enolase In Complex With An Analog Of The Natural Product Sf-2312 Metabolite.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2019-11-27 Classification: LYASE Ligands: TLA, SO4, MG, GOL, KVM |
 |
Staphylococcus Aureus Dihydrofolate Reductase In Complex With Nadph And Methotrexate
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2019-10-23 Classification: antimicrobial protein, oxidoreductase Ligands: MTX, NAP, GOL |
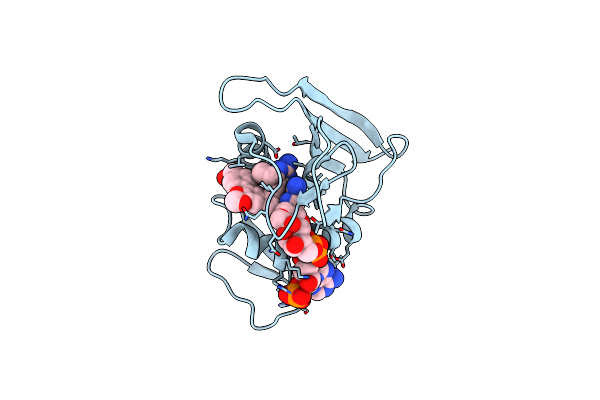 |
Staphylococcus Aureus Dihydrofolate Reductase In Complex With Nadph And Ucp1232
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-10-23 Classification: ANTIMICROBIAL PROTEIN Ligands: O71, XNP, NAP |
 |
Crystal Structure Of Enolase From Escherichia Coli With Bound 2-Phosphoglycerate Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2018-10-31 Classification: LYASE Ligands: 2PG, MG, SO4, GOL |
 |
Crystal Structure Of Enolase From E. Coli With A Mixture Of Apo Form, Substrate, And Product Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-10-31 Classification: LYASE Ligands: MG, SO4, GOL, PEP, 2PG, MES |
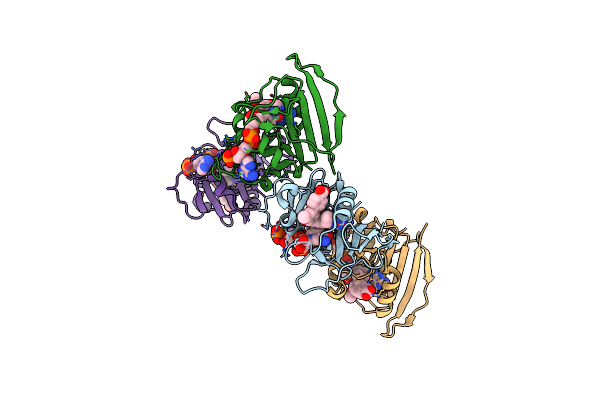 |
Mycobacterium Tuberculosis Dihydrofolate Reductase Complexed With Beta-Nadph And 3'-[(2R)-4-(2,4-Diamino-6-Ethylpyrimidin-5-Yl)But-3-Yn-2-Yl]-5'-Methoxy[1,1'-Biphenyl]-4-Carboxylic Acid
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-05-23 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: NDP, G6Y |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase Complexed With Beta-Nadph And 4-[3-[3-[2,4-Bis(Azanyl)-6-Ethyl-Pyrimidin-5-Yl]Prop-2-Ynyl]-5-Methoxy-Phenyl]Benzoic Acid
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2018-05-23 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: NDP, U75, ACT, NA, GOL |
 |
Mycobacterium Tuberculosis Dihydrofolate Reductase Complexed With Beta-Nadph And N-(4-{[(2-Amino-4-Oxo-3,4-Dihydropteridin-6-Yl)Methyl]Amino}-2-Hydroxybenzene-1-Carbonyl)-L-Glutamic Acid
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-05-23 Classification: oxidoreductase/oxidoreductase inhibitor Ligands: NDP, G7D, GOL, SO4 |

