Search Count: 23
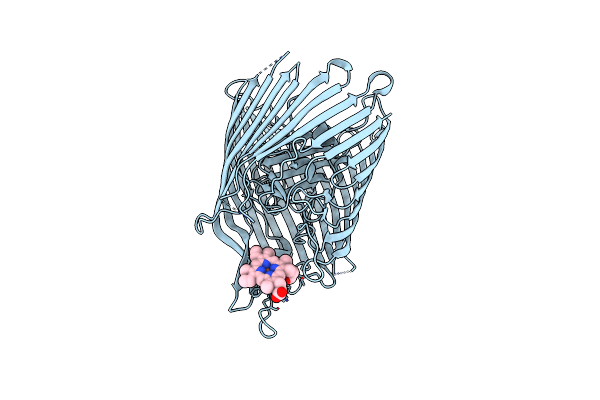 |
The Crystal Structure On The Heme/Hemoglobin Transporter Chua, In Complex With Heme
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN Ligands: HEM |
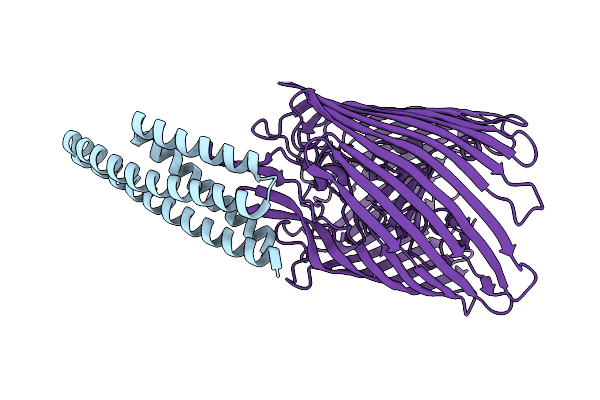 |
Cryo-Em Structure Of The Heme/Hemoglobin Transporter Chua, In Complex With De Novo Designed Binder G7
Organism: Synthetic construct, Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
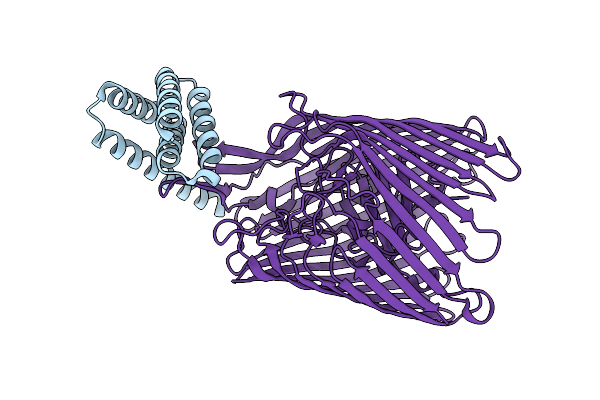 |
Cryo-Em Structure Of The Heme/Hemoglobin Transporter Chua, In Complex With De Novo Designed Binder H3
Organism: Synthetic construct, Escherichia coli cft073
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN |
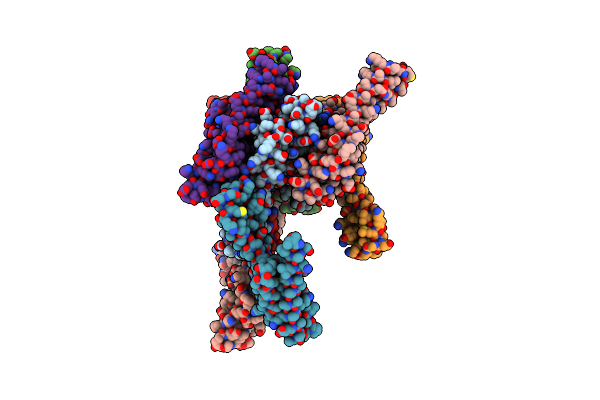 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2025-05-21 Classification: TRANSPORT PROTEIN Ligands: HOH |
 |
The Cryoem Structure Of The High Affinity Carbon Monoxide Dehydrogenase From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: CUN, MCN, FAD, FES |
 |
The Crystal Structure Of Coxg From M. Smegmatis, Minus Lipid Anchoring C-Terminus.
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-10-02 Classification: ELECTRON TRANSPORT |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2023-10-25 Classification: UNKNOWN FUNCTION Ligands: GOL, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN |
 |
Growth Factor Receptor-Bound Protein 2 (Grb2) Bound To Phosphorylated Peak3 (Py24) Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-06-07 Classification: SIGNALING PROTEIN Ligands: EDO, SO4 |
 |
Crystal Structure Of The N-Terminal Domain Of The Cryptic Surface Protein (Cd630_25440) From Clostridium Difficile.
Organism: Clostridioides difficile 630
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-05-10 Classification: UNKNOWN FUNCTION Ligands: CL, EDO, GOL |
 |
The 2.19-Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Complex Minus Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.67 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, OH, MG, VK3, F3S |
 |
The Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Full Complex Focused Refinement Of Stalk
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: OXIDOREDUCTASE Ligands: 3NI, FCO, MG, MQ9, F3S |
 |
The 1.52 Angstrom Cryoem Structure Of The [Nife]-Hydrogenase Huc From Mycobacterium Smegmatis - Catalytic Dimer (Huc2S2L)
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2023-01-04 Classification: ELECTRON TRANSPORT Ligands: O, MG, FCO, 3NI, VK3, F3S |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: X7Y |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: X8J |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-09-22 Classification: TRANSFERASE Ligands: XBD, PEG, PG4 |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-02 Classification: LIPID BINDING PROTEIN Ligands: ZN |

