Search Count: 94
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME Ligands: MG, NAD, SPM, SRY, K, ZN, FES, ATP, GNP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME Ligands: NAD, MG, K, ZN, FES, ATP, GDP, SAH |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME Ligands: NAD, SPM, SRY, MG, K, ZN, FES, ATP, GNP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME Ligands: NAD, SPM, SRY, MG, K, ZN, FES, ATP, GNP, FME, GTP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: RIBOSOME Ligands: ZN, MG, PNS |
 |
Organism: Saccharomyces cerevisiae (strain fostersb)
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2019-04-24 Classification: STRUCTURAL PROTEIN |
 |
Organism: Carboxydothermus hydrogenoformans (strain atcc baa-161 / dsm 6008 / z-2901), Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2019-04-24 Classification: STRUCTURAL PROTEIN Ligands: CA |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Chlorobaculum tepidum (strain atcc 49652 / dsm 12025 / nbrc 103806 / tls)
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2019-04-24 Classification: STRUCTURAL PROTEIN Ligands: MPD |
 |
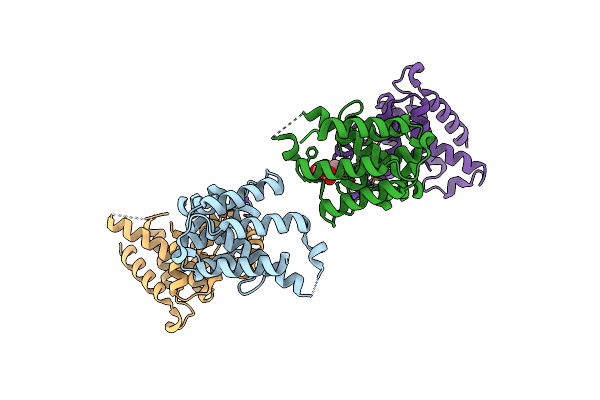
2.55 Angstrom Resolution Structure Of 3-Phosphoshikimate 1-Carboxyvinyltransferase (Aroa) From Coxiella Burnetii In Complex With Shikimate-3-Phosphate, Phosphate, And Potassium
Organism: Coxiella burnetii (strain rsa 493 / nine mile phase i)
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2015-05-13 Classification: TRANSFERASE Ligands: S3P, PO4, K, BME |
 |
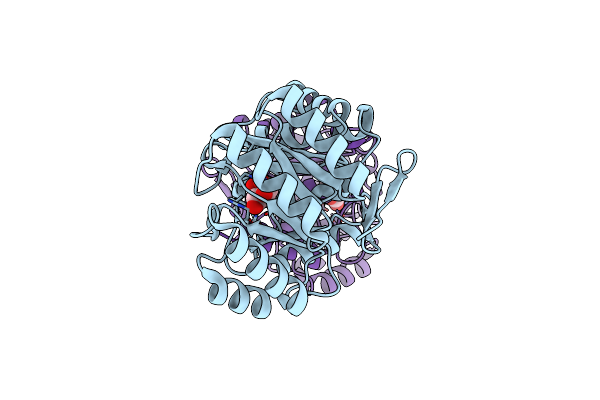
1.85 Angstrom Crystal Structure Of Lmo0812 From Listeria Monocytogenes Egd-E
Organism: Listeria monocytogenes serovar 1/2a
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-03-04 Classification: HYDROLASE |
 |
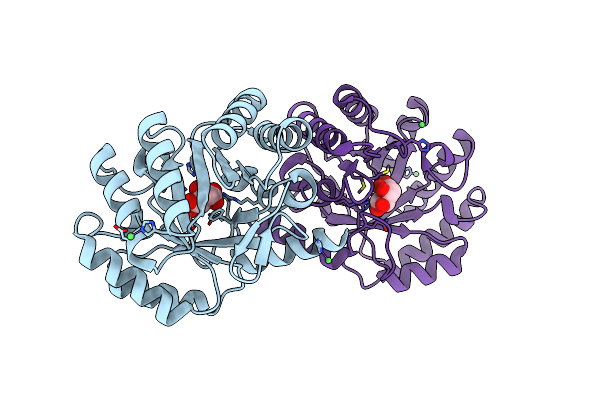
1.8 Angstrom Crystal Structure Of The Salmonella Enterica 3-Dehydroquinate Dehydratase (Arod) K170M Mutant In Complex With Quinate
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-01-30 Classification: LYASE Ligands: QIC |
 |
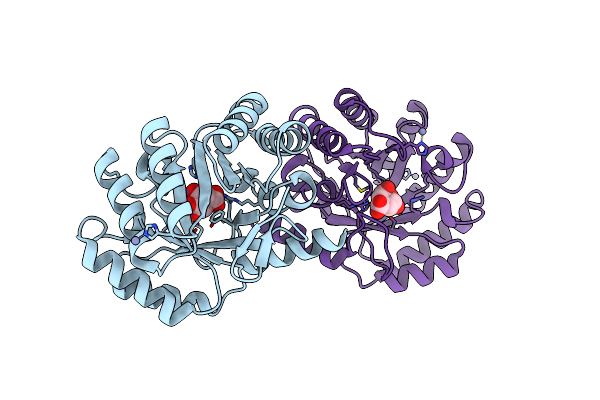
1.78 Angstrom Crystal Structure Of The Salmonella Enterica 3-Dehydroquinate Dehydratase (Arod) In Complex With Quinate
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2012-09-12 Classification: LYASE Ligands: QIC, NI |
 |
1.50 Angstrom Crystal Structure Of The Salmonella Enterica 3-Dehydroquinate Dehydratase (Arod) In Complex With Shikimate
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-09-12 Classification: LYASE Ligands: SKM, ZN |
 |
2.7 Angstrom Resolution Structure Of 3-Phosphoshikimate 1-Carboxyvinyltransferase (Aroa) From Coxiella Burnetii In A Second Conformational State
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-08-15 Classification: TRANSFERASE Ligands: BME |
 |
1.8 Angstrom Crystal Structure Of The 3-Dehydroquinate Dehydratase (Arod) From Salmonella Typhimurium Lt2 With Nickel Bound At Active Site
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-08-15 Classification: LYASE Ligands: NI, SIN |
 |
2.50 Angstrom Resolution Structure Of 3-Phosphoshikimate 1-Carboxyvinyltransferase (Aroa) From Coxiella Burnetii In Complex With Phosphoenolpyruvate
Organism: Coxiella burnetii
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-11 Classification: TRANSFERASE Ligands: PEP, SO4 |
 |
2.5 Angstrom Resolution Crystal Structure Of Bifidobacterium Longum Chorismate Synthase
Organism: Bifidobacterium longum subsp. infantis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-04-04 Classification: LYASE Ligands: CL |

