Search Count: 45
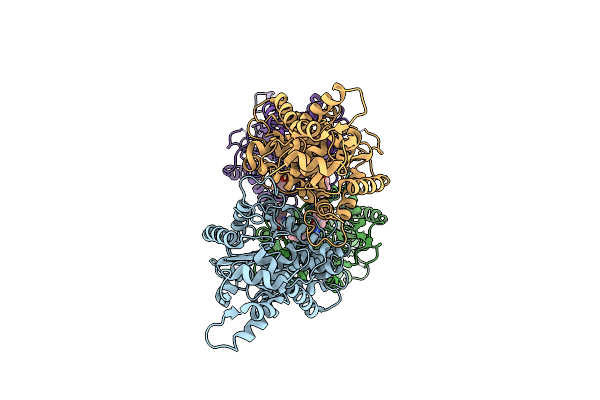 |
Room Temperature Serial Crystal Structure Of Glutaminase C In Complex With Inhibitor Upgl-00004
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-05-25 Classification: HYDROLASE/INHIBITOR Ligands: B4A |
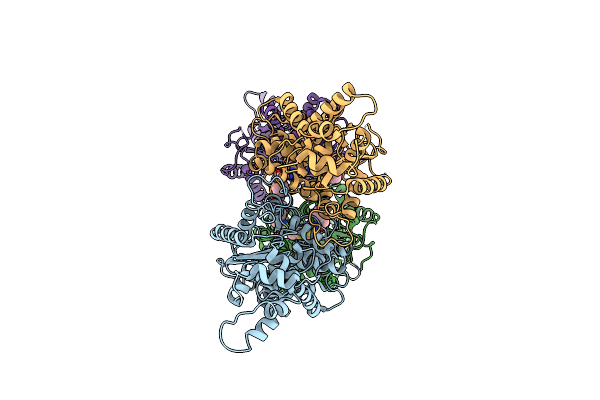 |
Room Temperature Serial Crystal Structure Of Glutaminase C In Complex With Inhibitor Bptes
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-05-25 Classification: HYDROLASE/INHIBITOR Ligands: 04A |
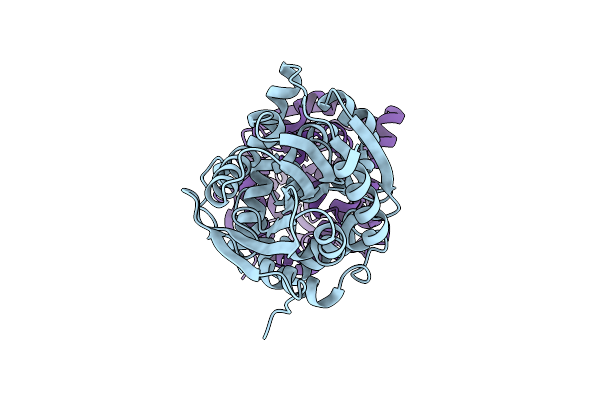 |
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 1 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE |
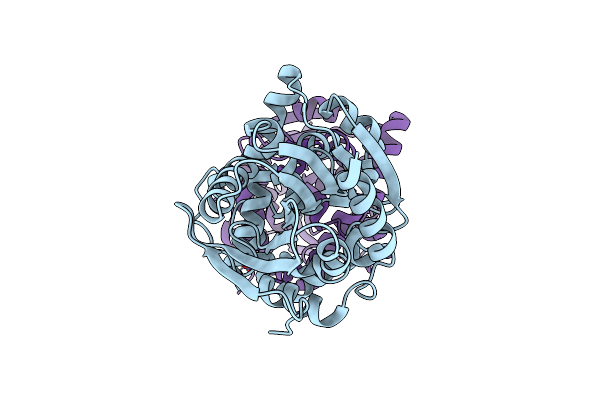 |
Fluoroacetate Dehalogenase, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Lysozyme, Room Temperature Structure Solved By Serial 3 Degree Oscillation Crystallography
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CL, NA |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure Solved By Serial 5Degree Oscillation Crystallography
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-27 Classification: TRANSPORT PROTEIN Ligands: HEM, CMO, SO4 |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using First 1 Degree Of Total 3 Degree Oscillation
Organism: Rhodopseudomonas palustris cga009
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Fluoroacetate Dehalogenase, Room Temperature Structure, Using Last 1 Degree Of Total 3 Degree Oscillation And 144 Kgy Dose
Organism: Rhodopseudomonas palustris (strain atcc baa-98 / cga009)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-03-27 Classification: HYDROLASE Ligands: CA |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure, First 2 Degrees Of 5 Degree Total Oscillation
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-03-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4 |
 |
Co-Bound Sperm Whale Myoglobin, Room Temperature Structure, Last 2 Degrees Of 5 Degree Total Oscillation And 160 Kgy Dose
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-03-27 Classification: OXYGEN TRANSPORT Ligands: HEM, CMO, SO4 |
 |
Crystal Structure Of Adp Ribosyl Cyclase Complexed With A Substrate Analog And A Product Nicotinamide
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: NFD, AVV, NCA |
 |
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: NAD |
 |
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: N1C |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: AVU |
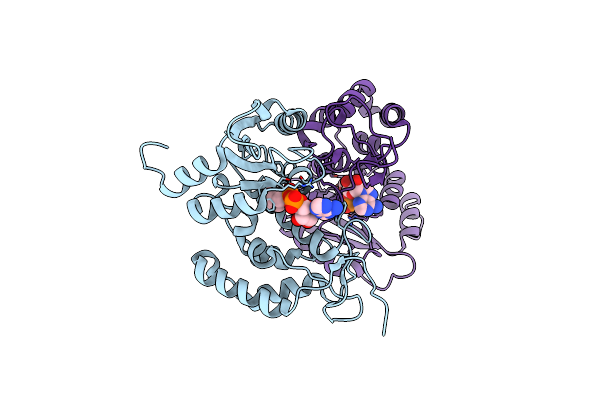 |
Crystal Structure Of Human Cd38 Complexed With An Analog Ribo-2'F-Adp Ribose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: AVW |
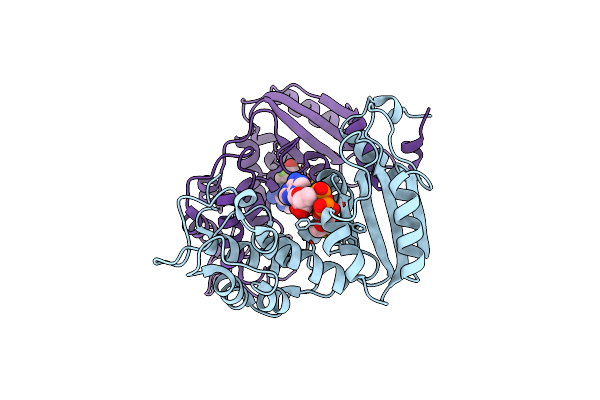 |
Crystal Structure Of Adp Ribosyl Cyclase Complexed With Ribo-2'F-Adp Ribose
Organism: Aplysia californica
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-07-28 Classification: HYDROLASE Ligands: AVW |
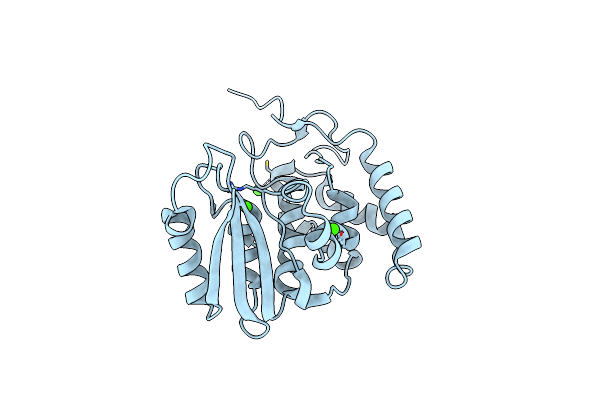 |
Conformational Closure Of The Catalytic Site Of Human Cd38 Induced By Calcium
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2008-11-18 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of Human Cd38 Extracellular Domain Complexed With A Covalent Intermediate, Ara-F-Ribose-5'-Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2008-11-04 Classification: HYDROLASE Ligands: RF5 |
 |
Crystal Structure Of Human Cd38 Extracellular Domain, Ara-F-Ribose-5'-Phosphate/Nicotinamide Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-11-04 Classification: HYDROLASE Ligands: RF5, NCA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2008-11-04 Classification: HYDROLASE Ligands: GTP |

