Search Count: 49
 |
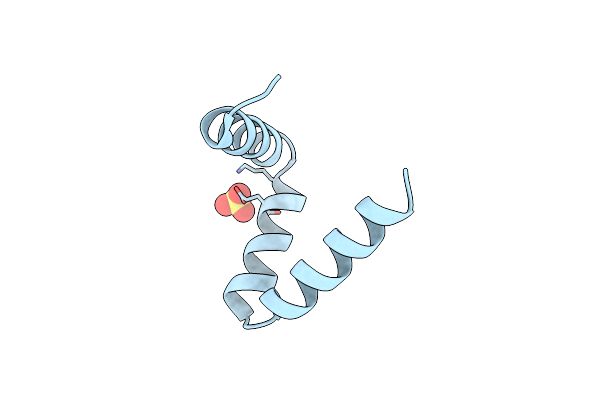
Solution Nmr Structure Of A Peptide Encompassing Residues 2-19 Of The Human Formin Inf2
|
 |
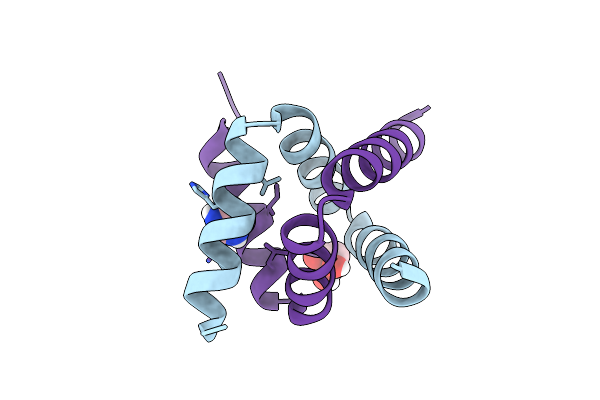
Solution Nmr Structure Of A Peptide Encompassing Residues 2-36 Of The Human Formin Inf2
|
 |
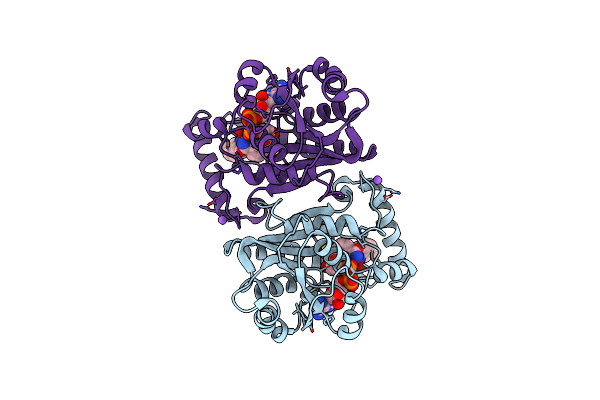
Crystal Structure Of An Enoyl-[Acyl-Carrier-Protein] Reductase Inha From Mycobacterium Fortuitum Bound To Nad And Nitd-916
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: OXIDOREDUCTASE Ligands: NAD, 3KY, NA, CL |
 |
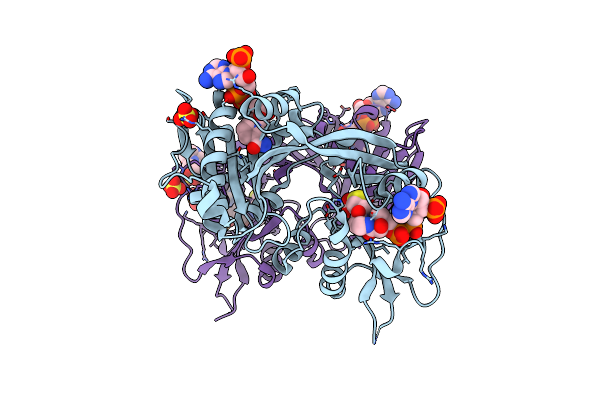
Crystal Structure Of A Enoyl-[Acyl-Carrier-Protein] Reductase (Inha) From Mycobacterium Abscessus Bound To Nad And Nitd-916
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-10-05 Classification: OXIDOREDUCTASE Ligands: NAD, 3KY, NA |
 |
Crystal Structure Of Mab_4324 A Tandem Repeat Gnat From Mycobacterium Abscessus
Organism: Mycobacteroides abscessus (strain atcc 19977 / dsm 44196 / cip 104536 / jcm 13569 / nctc 13031 / tmc 1543)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-05-04 Classification: TRANSFERASE Ligands: NDP, COA, ACO, ACT, SO4, NAI, EPE |
 |
Crystal Structure Of The M. Abscessus Leurs Editing Domain In Complex With Epetraborole-Amp Adduct
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: SO4 |
 |
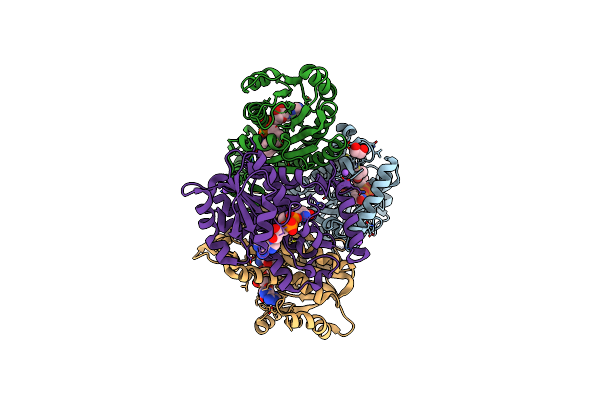
Crystal Structure Of The M. Abscessus Leurs Editing Domain In Complex With Epetraborole-Amp Adduct
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-10-06 Classification: LIGASE/LIGASE INHIBITOR Ligands: 365, SO4 |
 |
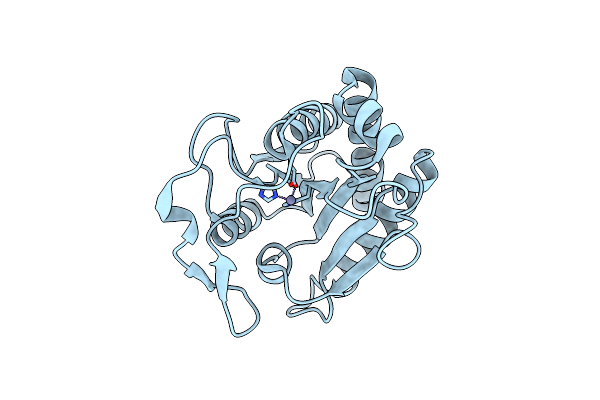
Crystal Structure Of Enoyl-[Acyl-Carrier-Protein] Reductase Inha From Mycobacterium Abscessus In Complex With Nad
Organism: Mycobacteroides abscessus (strain atcc 19977 / dsm 44196 / cip 104536 / jcm 13569 / nctc 13031 / tmc 1543)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-01-13 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, NA |
 |
Organism: Drosophila x virus (isolate chung/1996)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-18 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Drosophila x virus (isolate chung/1996)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-11-18 Classification: VIRAL PROTEIN Ligands: IMD, GOL |
 |
Crystal Structure Of The Apo Form Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Abscessus.
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Smegmatis
Organism: Mycolicibacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The N-Acetylmuramyl-L-Alanine Amidase, Ami1, From Mycobacterium Abscessus Bound To L-Alanine-D-Isoglutamine
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-18 Classification: SUGAR BINDING PROTEIN Ligands: ZGL, ALA, ZN |
 |
Crystal Structure Of Enoyl-[Acyl-Carrier-Protein] Reductase [Nadh] (Inha) From Mycobacterium Abscessus
Organism: Mycobacteroides abscessus (strain atcc 19977 / dsm 44196 / cip 104536 / jcm 13569 / nctc 13031 / tmc 1543)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-11-11 Classification: OXIDOREDUCTASE Ligands: EDO, SO4 |
 |
Structure Of Enoyl-[Acyl-Carrier-Protein] Reductase [Nadh] From Mycobacterium Fortuitum Bound To Nad
Organism: Mycolicibacterium fortuitum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-10-28 Classification: OXIDOREDUCTASE Ligands: NAD, EDO, ACT |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2020-09-09 Classification: TRANSFERASE Ligands: ACO, SO4 |
 |
Crystal Structure Of The Mycobacterial Trehalose Monomycolate Transport Factor A, Ttfa
Organism: Mycolicibacterium smegmatis mc2 155
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-12-25 Classification: LIPID TRANSPORT Ligands: SO4 |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-10 Classification: ANTIBIOTIC Ligands: ACO |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-07-10 Classification: ANTIBIOTIC Ligands: CIT, P4K, ACT |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-07-10 Classification: ANTIBIOTIC Ligands: BTB, SO4 |

