Search Count: 22
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 7.5, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLA, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLI, IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 5.6, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 5.6, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLA, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: MLI, IOD |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 9, Room Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structure Of Deuterated Gamma-Chymotrypsin At Ph 9, Cryo Temperature
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: SO4, IOD |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: IOD, SO4 |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Catalytic Core
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-04-25 Classification: LYASE Ligands: PLP, ACT, CA, NA, CL, PEG, PGE, EDO |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Plp-L-Serine Intermediate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2018-04-25 Classification: LYASE Ligands: EVM, CA, NA, CL, PEG, PGE, EDO |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Plp-Aminoacrylate Intermediate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2018-04-25 Classification: LYASE Ligands: P1T, CA, NA, CL, PEG, PGE, EDO |
 |
Crystal Structures Of Cystathionine Beta-Synthase From Saccharomyces Cerevisiae: The Structure Of The Pmp Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-04-25 Classification: LYASE Ligands: PMP, CA, NA, CL, PEG, EDO |
 |
Benzaldehyde Dehydrogenase, A Class 3 Aldehyde Dehydrogenase, With Bound Nadp+ And Benzoate Adduct
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2017-04-12 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
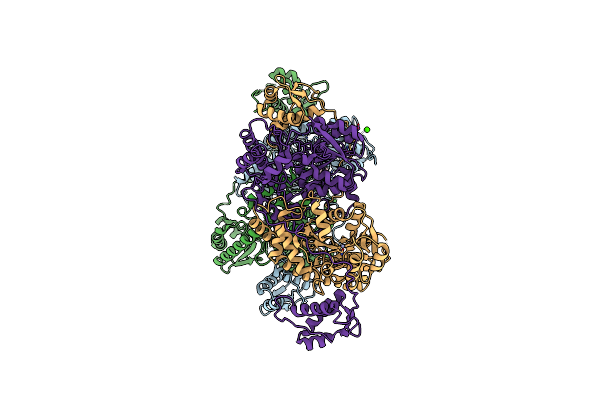
The Crystal Structure Of Bacillus Subtilis Gabr, An Autorepressor And Plp- And Gaba-Dependent Transcriptional Activator Of Gabt
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION REGULATOR Ligands: ZN, IMD, ACT |
 |
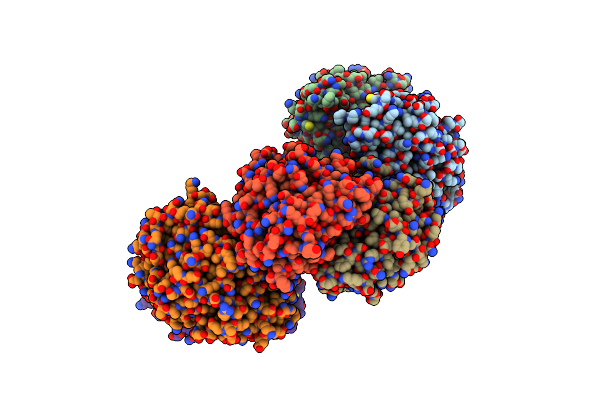
Crystal Structure Of Bacillus Subtilis Gabr, An Autorepressor And Transcriptional Activator Of Gabt
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION ACTIVATOR Ligands: ACE, ZN, CA |
 |
Organism: Naegleria gruberi
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2013-10-02 Classification: TRANSFERASE |

