Search Count: 18
 |
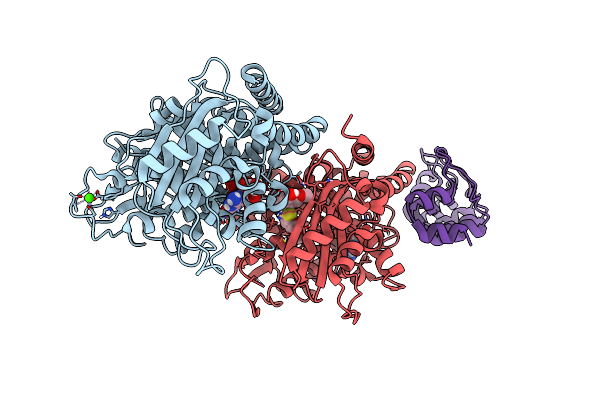
Photostatin (Photoswitchable Azo-Combretastatin) Z-Pst27 Bound To Tubulin-Darpin D1 Complex
Organism: Bos taurus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-09-04 Classification: CELL CYCLE Ligands: GTP, MG, CA, A1IBN |
 |
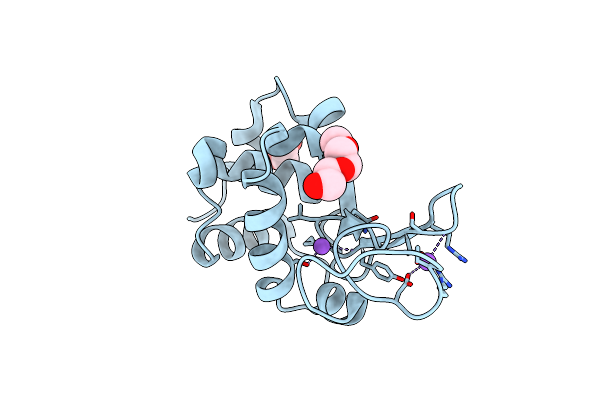
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-28 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, I8R |
 |
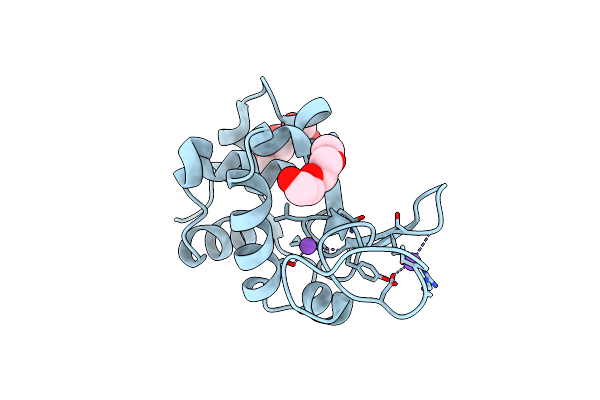
Organism: Asplenium bulbiferum subsp. bulbiferum
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: EDO, CL, NA, 1PE |
 |
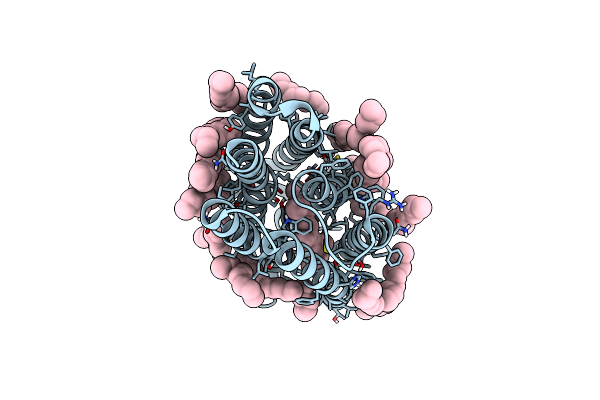
Organism: Asplenium bulbiferum subsp. bulbiferum
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-12-13 Classification: HYDROLASE Ligands: CL, NA, 1PE, EDO |
 |
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2023-12-13 Classification: MEMBRANE PROTEIN Ligands: RET, LFA |
 |
Krokinobacter Eikastus Rhodopsin 2 (Kr2) Extrapolated Map 1Us After Light Activation
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-13 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, LOC, ACP |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, ACP |
 |
Organism: Mus musculus, Pavo cristatus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, BKF, ACP |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, VLB, GDP |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, V1O, ACP |
 |
Organism: Synthetic construct, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, EP, LOC, GDP, ACP |
 |
Drug Cocktail (Colchicine, Epothilone A, Peloruside, Ansamitocin P3, Vinblastine) Bound To Tubulin (T2R-Ttl) Complex
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, LOC, EP, POU, VLB, BKF, ACP |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-05-30 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2018-05-30 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
 |

