Search Count: 5
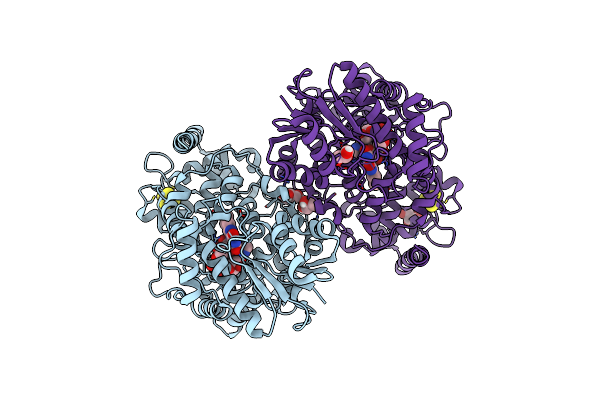 |
Crystal Structure Of Iron-Sulfur Cluster Containing Photolyase Phrb Mutant I51W
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-01-11 Classification: LYASE Ligands: FAD, SF4, GOL |
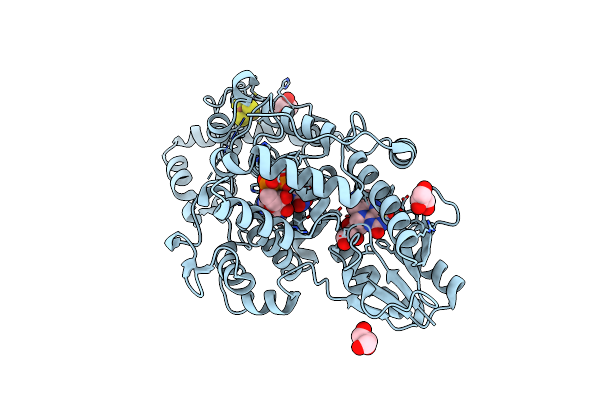 |
Crystal Structure Of Iron-Sulfur Cluster Containing Bacterial (6-4) Photolyase Phrb - Y424F Mutant With Impaired Dna Repair Activity
Organism: Agrobacterium fabrum (strain c58 / atcc 33970)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-01-11 Classification: LYASE Ligands: FAD, DLZ, SF4, GOL |
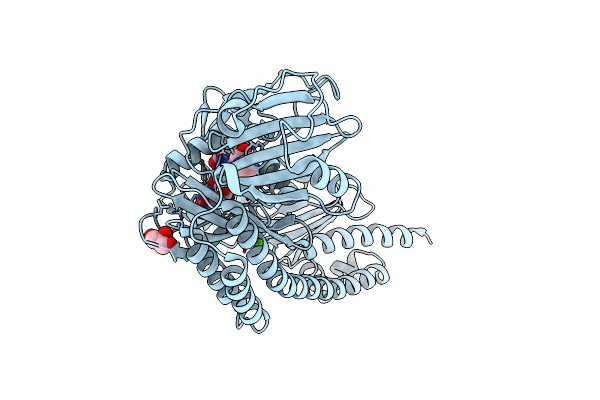 |
The Surface Engineered Photosensory Module (Pas-Gaf-Phy) Of The Bacterial Phytochrome Agp1 (Atbphp1) In The Pr Form, Chromophore Modelled With An Endocyclic Double Bond In Pyrrole Ring A.
Organism: Agrobacterium fabrum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-08-03 Classification: SIGNALING PROTEIN Ligands: BLA, CA, GOL, IMD |
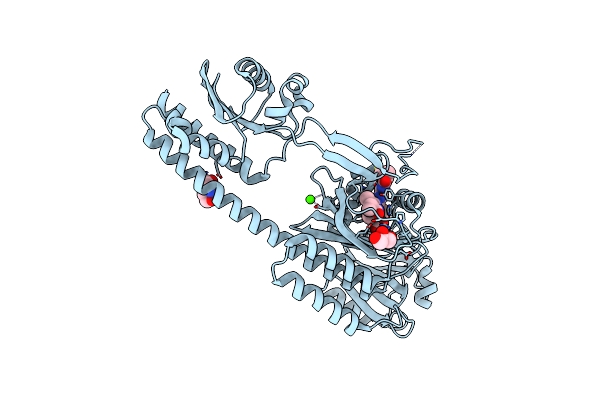 |
The Photosensory Module (Pas-Gaf-Phy) Of The Bacterial Phytochrome Agp1 (Atbphp1) In The Pr Form, Chromophore Modelled With An Endocyclic Double Bond In Pyrrole Ring A
Organism: Agrobacterium fabrum str. c58
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-08-03 Classification: SIGNALING PROTEIN Ligands: BLA, CA, TAM, GOL |
 |
Crystal Structure Of A Bacterial Class Iii Photolyase From Agrobacterium Tumefaciens At 1.67A Resolution
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2015-03-25 Classification: LYASE Ligands: MHF, FAD, SO4, TRS |

